Evaluation of five selective media for the detection of Pseudomonas aeruginosa using a strain panel...
Transcript of Evaluation of five selective media for the detection of Pseudomonas aeruginosa using a strain panel...
Journal of Microbiological Methods 99 (2014) 8–14
Contents lists available at ScienceDirect
Journal of Microbiological Methods
j ourna l homepage: www.e lsev ie r .com/ locate / jmicmeth
Evaluation of five selective media for the detection of Pseudomonasaeruginosa using a strain panel from clinical, environmental andindustrial sources
Rebecca Weiser a,⁎, Denise Donoghue b, Andrew Weightman a, Eshwar Mahenthiralingam a
a Cardiff School of Biosciences, Cardiff University, Cardiff, UKb Unilever Research & Development, Port Sunlight, Wirral, UK
⁎ Corresponding author at: Cardiff School of BiosciencCardiff University, Museum Avenue, Cardiff CF10 3AT, UK
E-mail address: [email protected] (R. Weiser).
0167-7012/$ – see front matter © 2014 Elsevier B.V. All rihttp://dx.doi.org/10.1016/j.mimet.2014.01.010
a b s t r a c t
a r t i c l e i n f oArticle history:Received 1 November 2013Received in revised form 17 January 2014Accepted 17 January 2014Available online 25 January 2014
Keywords:Culture-dependent detectionPseudomonas aeruginosaSelective media
Isolation and correct identification of the opportunistic pathogen and industrial contaminant Pseudomonasaeruginosa are very important and numerous selective media are available for this purpose. A novel comparisonof five selective media having positive (acetamide-based agars), negative (Pseudomonas CN selective agar[Oxoid Ltd.] and Pseudomonas Isolation agar [Sigma-Aldrich Company Ltd.]) and chromogenic (chromID®P. aeruginosa [bioMérieux]) selection strategies was performed using a systematically designed bacterialtest panel (58 P. aeruginosa and 90 non-P. aeruginosa strains including those commonly misidentified asP. aeruginosa by culture-dependent techniques). Standardised inocula were added to the selective media andthe results were recorded after 24 and 72 h. After 72 h of incubation at 37 °C chromID® P. aeruginosa displayedthe highest specificity (70%) and had good sensitivity (95%), although the sensitivity was negatively impacted bythe large variation in colour of P. aeruginosa colonies, which hampered interpretation. Both media containinginhibitory selective agents performed very similarly, both having 100% sensitivity and a specificity of approxi-mately 30%. Raising the incubation temperature to 42 °C increased the specificity of Pseudomonas CN selectiveagar and Pseudomonas isolation agar (61% and 47% respectively after 72 h), but increased the number of falsepositives encountered with the chromogenic medium, decreasing its specificity to 68% after 72 h. Growth onthe acetamide agars was weak for all strains and it was often difficult to determine whether true growth hadoccurred. This, compounded by the low specificity of the acetamide agars (b26%), suggested they were lesssuitable for application to clinical or industrial settings without further modification. Overall, the chromogenicagar was the most selective but further consideration is required to optimise interpretation of results.
© 2014 Elsevier B.V. All rights reserved.
1. Introduction
Pseudomonas aeruginosa is an extremely versatile microorganismwith minimal growth requirements and the ability to survive in diversehabitats (Morrison andWenzel, 1984; Stover et al., 2000). As an oppor-tunistic pathogen, P. aeruginosa can cause a broad spectrum of illness in-cluding pneumonia, urinary tract infections, wound and burn infections,bacteraemia and septicaemia (Kerr and Snelling, 2009) in susceptiblehosts. Notably, P. aeruginosa is a major pathogen associated with cysticfibrosis (CF) patients and is responsible for chronic, life-threatening pul-monary infections which are extremely difficult to eradicate (Gomezand Prince, 2007). In addition to being clinically relevant, P. aeruginosaalso impacts on industry as a commonly encountered contaminant of in-dustrial processes and products. Due to the health risks posed byP. aeruginosa, it is widely considered an ‘objectionable organism’ to befound in consumer products (Jimenez, 2007; Sutton and Jimenez,
es, Main Building, Room 0.20B,. Tel.: +44 2920 874 648.
ghts reserved.
2012), and represents a serious concern for manufacturers. Relativelylittle is known, however, about P. aeruginosa in industrial settings.
Correct identification of P. aeruginosa is paramount and themajorityof routine diagnostic tests for P. aeruginosa are reliant on culture-dependent methods (Deschaght et al., 2011). Numerous selectiveagars are available for the isolation of P. aeruginosa including thosethat include inhibitory selective agents such as antibiotics and biocides(Lilly and Lowbury, 1972), those that use positive selection (Szitaet al., 1998) and those that use chromogenic detection (Laine et al.,2009). Cetrimide-containing isolation media are commonly used bothin clinical and industrial settings (FDA, 1998) but can lack selectivity(Kodaka et al., 2003; Laine et al., 2009). These media are often also de-signed to enhance pigment production as synthesis of pyocyanin canunequivocally identify P. aeruginosa (Reyes et al., 1981). Only 90–95%of isolates produce pyocyanin (Smirnov and Kiprianova, 1990), howev-er, which can confound identification. Media containing acetamide asthe positive selective agent have not been evaluated extensively, al-though have been reported to have good selectivity and may be usefulfor detecting low numbers of cells from ‘stressed’ environments (Szitaet al., 2007; Szita et al., 1998). In these media acetamide represents
9R. Weiser et al. / Journal of Microbiological Methods 99 (2014) 8–14
the sole carbon and nitrogen source, which few species other thanP. aeruginosa can dissociate to release ammonia and acetic acid (Szitaet al., 1998). Auxotrophic P. aeruginosa isolates, such as those encoun-tered in respiratory infections in cystic fibrosis patients (Thomas et al.,2000), have difficulty growing on minimal media meaning that furthermodification of the acetamide media may be necessary to improvesensitivity. Only one study has evaluated the recently developedchromID® P. aeruginosa (manufactured by bioMérieux, La Balme-les-Grottes, France) showing the chromogenic agar to have higher selectiv-ity than the Pseudomonas CN selective agar (Oxoid, Basingstoke, UK).By virtue of an unambiguous colour change the medium has demon-strated potential for the simultaneous isolation and identification ofP. aeruginosa (Laine et al., 2009), warranting further assessment.
Unlike previous studies, which in general have considered four orfewer selective agars with similar selection strategies (Kodaka et al.,2003), we evaluated five selective media in parallel for the detectionof P. aeruginosa, including those using inhibitory selective agents, apositive selective agent and chromogenic detection methods (Table 1).In addition, rather than measuring the performance of these agars aspart of prospective surveillance for P. aeruginosa in environmental orclinical samples, a bacterial test panel was systematically assembled toinclude genetically distinct P. aeruginosa strains validated by molecularidentification and strains from commonly mistaken non-P. aeruginosaspecies (Spilker et al., 2004) andGram-negative pathogens encounteredin CF patients (Hauser et al., 2011). The modified Z-agar (acetamideagar) differed from the original Z-agar (Szita et al., 1998) by the additionof sufficient nutrient supplementation to facilitate growth of auxotro-phic P. aeruginosa isolates that are commonly encountered in CF(Thomas et al., 2000). Finally, as well as including strains from clinicaland environmental sources, our comparative agar study is the first toinclude P. aeruginosa strains from industry as an emerging problemarea (Sutton and Jimenez, 2012). The efficacy of these isolation agarsacross strains, commonly mistaken species and accounting for strainsource is discussed.
2. Methods
2.1. Bacterial strains and culture conditions
The strain panel comprised 148 strains (summarised in Tables 2 and4), including 58 P. aeruginosa and 90 non-P. aeruginosa strains (fulldetails in Supplementary Table S1). All P. aeruginosa isolates wereconfirmed to be P. aeruginosa by the use of a species-specific PCR(oprL PCR; De Vos et al., 1997). The non-P. aeruginosa strains belongedto other Pseudomonas species, other Gram-negative species and Gram-positive species. Strains originated from clinical, environmental andindustrial sources. Eighty-eight strains were obtained from a collectionheld at Cardiff University, whilst 58 strains were supplied by industryor recovered from industrial sources, and 2 were isolated from thedomestic environment. All P. aeruginosa strains were routinely grownon Tryptone Soya Agar (TSA; Oxoid Ltd., Basingstoke, UK) or in TryptoneSoya Broth (TSB; Oxoid Ltd.) overnight (18 h) at 37 °C. All other isolateswere grown at 30 °C. Overnight cultures were prepared by inoculating3 ml of TSB with fresh (b72 h) growth material collected using a sterilepipette tip from an area of confluent growth on a pure culture plate, andincubated on an orbital shaker (150 rpm). Bacterial strains were storedfrozen in individual vials in TSB containing 8% dimethylsulphoxide(DMSO) at−80 °C.
2.2. Culture media
TSB, TSA, Pseudomonas CN selective agar (PCN; Oxoid Ltd.) and Pseu-domonas isolation agar (PIA; Sigma-Aldrich Company Ltd., Dorset, UK)were prepared according to the manufacturers' instructions. Glyceroland ethanol (analytical reagent grade) were supplied by Fisher ScientificUK Ltd. (Leicestershire, UK). The chromogenic medium chromID®
P. aeruginosa (chromID Pa) was obtained as pre-poured plates frombioMérieux UK Ltd. (Basingstoke, UK). Z-agar was prepared as describedby Szita et al. (1998), and modified Z-agar had the same formula asZ-agar, but with the addition of 0.01% (w/v) Bacto™ Casamino acids(BD UK Ltd., Oxford, UK) and 0.01% (w/v) yeast extract (Oxoid Ltd.).
2.3. Comparison of selective media
Overnight cultures of strains were diluted to an optical density (OD)of 1 at 600 nm (corresponding to approximately 108 cfu/ml) and 200 μlof the diluted culture pipetted into separate wells of a 96 well micro-titre plate. The cultures were replica plated, using a 48 pin replicator(approximately 105 cfu/ml transferred) from the 96 well plate ontoTSA, PCN, PIA, chromID Pa, Z-agar and modified Z-agar in triplicate. Allagar plates were incubated at 37 °C and 42 °C for 72 h, except the TSAplates for the non-P. aeruginosa panel which were incubated at 30 °Cfor 72 h, and the Z-agar andmodified Z-agar plates whichwere only in-cubated at 37 °C. Results were recorded at 24 and 72 h. Positive resultson chromID Pa were originally intended to be taken as growth accom-panied by a red/pink/purple colouration as described previously(Laine et al., 2009), but this interpretationwasmodified to also incorpo-rate strains that displayed a pink-brown colour. Pigment production(pyocyanin and fluorescein) by P. aeruginosa strains on TSA, PCN andPIA was recorded at 37 °C. Growth on any of the other media wastaken as a positive result, regardless of the presence/absence of pigmentproduction.
2.4. Evaluation criteria
The sensitivity and specificity of the agars were calculated. The sen-sitivity was taken as the number of P. aeruginosa strains identified aspositive for growth on the media, divided by the total number ofP. aeruginosa strains screened; the probability of a true positive. Thespecificity was taken as the number of non-P. aeruginosa strains nega-tive for growth on the media, divided by the total number of non-P. aeruginosa strains screened; the probability of a true negative(Lalkhen andMcCluskey, 2008). Positive and negative predictive values(PPV/NPV) were calculated to determine the proportions of true posi-tives and negatives for the study population. The PPV was taken as thenumber of P. aeruginosa strains identified as positive for growth onthe media, divided by the total number of strains (both P. aeruginosaand non-P. aeruginosa) positive for growth. The NPV was taken as thenumber of non-P. aeruginosa strains negative for growth on themedia, divided by the total number of strains (both P. aeruginosa andnon-P. aeruginosa) which were negative for growth (Lalkhen andMcCluskey, 2008).
3. Results
3.1. Sensitivity of the five media
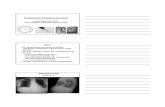
PCN, PIA and mZ-agar demonstrated 100% sensitivity as all 58P. aeruginosa strains grew on these agars after 24 h of incubation at37 °C (Table 2). The sensitivity of the chromogenic agar did not reachthis level due to the variable colours produced by certain P. aeruginosastrains (Fig. 1). Overall, 47 out of 58 strains of P. aeruginosa strainswere correctly identified after 24 h (81% sensitivity), with 5 strainsnot displaying adequate growth and 6 giving the wrong colour (yellow,orange, light brown, green and black), which may have reflected a defi-ciency in β-alanyl aminopeptidase activity. After 72 h the sensitivity ofchromID Pa increased to 95%. Growth on the Z-agars was muchweakerthan that on the other agars (Fig. 2) and 1 auxotrophic P. aeruginosastrain LESB58 (Winstanley et al., 2009) did not grow at all on the un-modified Z-agar. The modified Z-agar had nutrient supplementationsufficient for the cultivation of the LESB58 strain although growth wasstill very weak. There did not appear to be any major differences in
Table1
Selectiveag
arsforthede
tectionof
Pseu
domon
asaerugino
saev
alua
tedin
thisstud
y.
Selectiveag
arSe
lectiveag
ent(
conc
.g/l)
Commen
tPros
Cons
Supp
lier/Re
f.
Pseu
domon
asselective
CNag
ar(PCN
)Ce
trim
ide(0.2)na
lidixic
acid
(0.015
)Gen
eral
med
ium
fortheisolationof
P.aerugino
saCo
mmercially
available(eithe
ras
powde
r+
supp
lemen
tor
pre-po
ured
plates)an
dthereforemoresu
itab
leforus
ein
clinical/ind
ustrialsetting
sthan
agarsthat
have
tobe
prep
ared
in-hou
se.
Other
speciesha
vebe
ensh
ownto
grow
onthis
med
ium
includ
ingPseu
domon
asspp.,B
urkh
olderia
spp.
andAc
hrom
obacterxylosoxida
ns(p
ersona
lob
servations
andLaineet
al.,20
09)
Oxo
idLtd.
Pseu
domon
asisolation
agar
(PIA)
Triclosan(0.025
)Gen
eral
med
ium
fortheisolationof
P.aerugino
saCo
mmercially
availableas
apo
wde
r,us
esadiffe
rent
selective
agen
ttoPA
B.C-N.U
sedby
Jimen
ezet
al.(19
99)an
dJim
enez
etal.(20
00)to
detect
P.aerugino
sain
indu
strial
samples.
Other
Pseu
domon
asspeciesmay
grow
onthismed
ium
Sigm
a-Aldrich
chromID®
P.aerugino
sa(chrom
IDPa
)
Antibiotics
aan
dβ-alany
l-resorufamine
Chromog
enic
detectionof
P.aerugino
sa.
Colonies
turn
aviolet
colour,w
ithor
witho
utametallic
gold
shee
n,du
eto
hydrolysisof
thech
romog
enic
aminop
eptida
sesu
bstrate
β-alany
l-resorufamine.
Commercially
availableas
pre-po
ured
plates.H
ighe
rselectivity
than
PCN(Laine
etal.,20
09).
Other
speciesmay
grow
ontheag
aran
dprod
ucea
colourationsimila
rto
P.aerugino
sa,inc
luding
Burkho
lderia
cepa
ciacomplex
bacteria
and
P.fluo
rescen
s(Laine
etal.,20
09).In
addition
,this
med
ium
isex
pens
ive.
bioM
érieux
Z-ag
arAcetamide(4)
Positive
selectionof
P.aerugino
sa.Few
othe
rspeciescanus
eacetam
ideas
both
acarbon
andnitrog
ensource
Dev
elop
edforisolationof
low
numbe
rsof
P.aerugino
safrom
food
/water.Stressedisolates
may
revive
better
onamed
ium
containing
apo
sitive
selectiveag
ent.Re
ported
toha
vego
odselectivity(Szita
etal.,19
98)
Growth
appe
arsas
athin
film
andsometim
esha
rdto
detect,especially
inthecase
ofau
xotrop
hicisolates.
Will
have
tobe
mad
eup
in-hou
se,w
hich
may
bedifficu
ltin
clinical/ind
ustrialsetting
s.
Szitaet
al.(19
98)
Mod
ified
Z-ag
ar(m
Z-ag
ar)
Acetamide(4)
Mod
ified
toim
prov
ethegrow
than
dde
tectionof
auxo
trop
hicP.aerugino
saisolates
bythead
dition
of0.01
%(w
/v)
casaminoacidsan
d0.01
%(w
/v)ye
astex
tract
Sameas
acetam
ideag
arW
illha
veto
bemad
eup
inho
use,which
may
bedifficu
ltin
clinical/ind
ustrialsetting
s.Th
isstud
y
aSe
lectivean
tibiotic
agen
tsno
tdisclosedby
bioM
érieux
.
10 R. Weiser et al. / Journal of Microbiological Methods 99 (2014) 8–14
growth characteristics between clinical, environmental and industrialstrains apart from after 72 h of incubation on chromID Pa, the 3 strainswhich gave negative results were all from industrial sources. TheP. aeruginosa panel was also screened for growth on PCN, PIA andchromID Pa at 42 °C. Whilst there were no changes in the sensitivityof PCN or PIA at this increased temperature, slight improvements instrain colouration on chromID Pa were observed which led to anincreased number of strains recorded as positive: after 24 and 72 hthe sensitivity of chromID Pa was 98%.
3.2. Pigment production
Pigment production on TSA, PCN and PIA was variable (Table 3). OnTSA pyocyanin production was easily identified but as it was often diffi-cult to distinguish between pyocyanin and fluorescein on PCN and PIA,strainswere scored as positive or negative for production of either/bothpigments (Supplementary Fig. S2). Over 90% of strains produced apigment on all three agars after 24 h of incubation. PIA was the mostsuccessful at enhancing pigment production; 100% of strains wereobserved to produce pigment after 72 h of incubation. The majority ofstrains produced a light green/yellow pigment on PIA indicative of fluo-rescein rather than pyocyanin production.
3.3. Selectivity of the five media
The most selective of the 5 media was chromID Pa (Table 4), whichafter 72 h of incubation at 37 °C had a specificity of −70%. Strains ofBurkholderia (n = 15), other Pseudomonas (n = 12) and Ochrobactrum(n = 1) species exhibited growth on chromID Pa that had a similarcolouration to strains of P. aeruginosa. Although at 24 h Z-agar had thesecond highest specificity of 42%, this dropped to 26% after 72 h. Itwas also very difficult to determine if true growth had occurred onboth of the Z-agars (Fig. 2) which resulted in a high proportion of posi-tive results being recorded asweak (+/−) rather than true positive. Theadditional nutrient supplementation in themodified Z-agar led to an in-crease in non-P. aeruginosa strains that could grow, and overall this agarwas the least selective of the 5, having a specificity of 6% after 72 hof incubation. The two agars containing inhibitory selective agents,PCN and PIA, performed very similarly having specificities of 30%and 23%, respectively, after 72 h of incubation. In general, strains ofAchromobacter (n = 2), Burkholderia (n = 28) and Pseudomonas (PIAn = 23; PCN n = 22) species were most likely to give false positiveson PCN and PIA.
Incubation at 42 °C for 72 h increased the specificities of PCN and PIAto 61% and 47%, respectively. These incubation conditions, however,also elevated the number of false positives recorded for chromID Pa,which decreased its specificity from 80% to 69% at 24 h and 70% to68% at 72 h. Of the 28 strains positive for growth at 37 °C on chromIDafter 24 h of incubation, 9 of these could not grow at 42 °C, but 19‘new’ false positives were observed (detailed in Table S1). The effect ofincreased temperature on the selectivity of Z-agar and modifiedZ-agar was not investigated due to the weak growth already seen at37 °C. Finally, it was observed that strains from different sources grewequally well on the 5 media tested in this study.
3.4. Presumptive identification of P. aeruginosa
The PPV andNPV values for the agars at 37 °C and 42 °C are given inTable S3. For this strain panel chromID Pa had the highest PPV at 37 °C(72% and 67% at 24 and 72 h, respectively) and at 42 °C after incubationfor 72 h (62%). Due to the increased numbers of chromID Pa false posi-tives at 42 °C, PCN was the agar with the highest PPV at this increasedtemperature after 24 h of incubation (68%). chromID Pa and Z-agarwere the only agars with a NPV less than 100% as not all P. aeruginosastrains were correctly identified on these media. The lowest NPV wasfor chromID after 24 h of incubation at 37 °C (87%), although this
Table 2Growth of Pseudomonas aeruginosa strains on selective media at 37 °C.
Species Source No. ofstrains
No. of strains exhibiting growth
PCNa PIAb chromID Pac Z-agar mZ-agard
24 h 72 h 24 h 72 h 24 h 72 h 24 h 72 h 24 h 72 h
P. aeruginosa CLIN 22 22 22 22 22 18 22 21 21 22 22ENV 6 6 6 6 6 5 6 6 6 6 6IND 30 30 30 30 30 24 27 30 30 30 30
Total 58 58 58 58 58 47 55 57 (3 +/−)e 57 (1 +/−) 58 (2 +/−) 58 (1 +/−)Sensitivity (%) 100 100 100 100 81 95 98 98 100 100
CLIN, clinical; ENV, environmental; IND, industrial.a PCN, Pseudomonas CN selective agar.b PIA, Pseudomonas isolation agar.c chromID Pa, chromID® P. aeruginosa; a positive result on chromID Pa was taken as growth accompanied by a red/pink/pink-brown/purple colouration.d mZ-agar, modified Z-agar.e Weakly growing strains (+/−) were recorded as an overall positive and are indicated in the table.
11R. Weiser et al. / Journal of Microbiological Methods 99 (2014) 8–14
increased to 95% after 72 h of incubation and 98% at 42 °C regardless ofincubation duration.
4. Discussion
There have beenmany selectivemedia developed for the isolation ofP. aeruginosa from clinical and environmental samples. The majority ofmedia currently available for the isolation of P. aeruginosa were devel-oped some decades ago and routinely used media, such as thosecontaining cetrimide, lack selectivity (Kodaka et al., 2003). In addition,despite the fact that the majority of diagnostics are still based onculture-dependent methods, not many novel media have been recentlydeveloped and few studies have systematically evaluated the perfor-mance of available media.
Fig. 1. Variable colony morphology of Pseudomonas aeruginosa strains on chromID® P. aerug(2a, 2b and 2c) of incubation at 37 °C. Growth accompanied by a red, pink, pink-brown or purpof other colours including yellow (1a top row right), orange (1a top rowmiddle), brown (not shoof chromID® P. aeruginosa increased after 72 h of incubation.
The strain collection used in this study was drawn from clinical andenvironmental sources, in addition to including strains from industrialcontamination incidents. It was the first study to include industrialstrains, which represented just under half of the P. aeruginosa andapproximately a third of the non-P. aeruginosa sub-panels. No majordifferences were observed, however, in the growth characteristics ofthe industrial strains compared to the rest of the panel. The choice ofpanel strains perhaps reflected the low levels of specificity and PPVvalues observedwith the PCN and PIA agars as strains of Achromobacter,Burkholderia and Pseudomonas species are known to have broad antimi-crobial resistance (Bador et al., 2011; Hogardt et al., 2009; Li andNikaido, 2009; Rose et al., 2009), and have previously been shown togrow on PCN (Kodaka et al., 2003), although no studies have evaluatedPIA. PCN and PIA are currently widely available commercially andperformed very similarly both in terms of sensitivity and specificity.
inosa. Results for 27 P. aeruginosa strains are shown after 24 h (1a, 1b and 1c) and 72 hle colour was recorded as a positive result for P. aeruginosa. Strains grew to produce a rangewn in photos), green (1c top rowmiddle) and black (1c middle rowmiddle). The sensitivity
Fig. 2. Growth of Pseudomonas aeruginosa and non-P. aeruginosa strains on Z agar andmodified Z-agar. Results are shown for 9 P. aeruginosa strains (1a and 1b) and 21 non-P. aeruginosa strains (2a and 2b) after 72 h of incubation at 37 °C. Z-agar plates aremarkedwith red and are shown in panels 1a and 2a. Modified Z-agar plates are marked with blueand shown in panels 1b and 2b. Growth was weak for both P. aeruginosa and non-P. aeruginosa strains on the Z-agars and it was difficult to determine whether true growthhad occurred inmany cases (for example 1amiddle rowmiddle). The addition of extra nu-trient supplementation in the modified Z-agar not only encouraged the growth ofP. aeruginosa strains (compare 1a to 1b) but also led to an increase in the number ofnon-P. aeruginosa strains that could grow on the agar (compare 2a to 2b).
12 R. Weiser et al. / Journal of Microbiological Methods 99 (2014) 8–14
Both are also formulated to encourage pigment production, being basedon King's A medium (King et al., 1954).
Although on TSA the primary pigment being produced appeared tobe pyocyanin, with 97% of strains being pigmented, growth on PCN andPIA was largely accompanied by the production of a light green/yellowpigment more indicative of fluorescein. This pigment alone cannot posi-tively identify P. aeruginosa as all other fluorescent pseudomonads arecapable of fluorescein production (Leisinger and Margraff, 1979). Inaddition, non-pigmented strains of P. aeruginosa may resemble otherspecies that can grow on PCN and PIA. This highlights that althoughpigment production can guide identification, subsequent diagnostictests are required to obtain conclusive results. In clinical and industrialsettings, growth at 42 °C is often used to differentiate P. aeruginosafrom other bacterial species, although this growth characteristic is notexclusive to P. aeruginosa. Incubation at 42 °C was considered in thecurrent study and was found to increase the specificity and PPV of PCNand PIA. A variety of chromogenic media have been designed for thedetection of pathogens including P. aeruginosa. Through inclusion ofchromogenic substrates for an enzyme produced by the target species,
Table 3Pigment production by Pseudomonas aeruginosa strains at 37 °C.
Selective agar
Tryptone soya agarPseudomonas CN selective agarPseudomonas isolation agar
a Strains producing pyocyanin, fluorescein or both pigments.
chromogenic media aim to simplify the isolation and identification pro-cess by linking an unambiguous colour change with growth of a targetorganism (Perry and Freydière, 2007). chromID® P. aeruginosa wasdeveloped by bioMérieux for the detection of P. aeruginosa and hasbeen evaluated by one study (Laine et al., 2009) which found the medi-um to have higher selectivity than PCN. The present study also deter-mined chromID Pa to have superior selectivity but the lowest overallsensitivity. If results for chromID Pa were read after 24 h of incubationat 37 °C, as suggested by the manufacturer, there was a considerablerisk of false negatives, although after 72 h of incubation the sensitivitywas greatly improved. This corroborates what was found by Laine et al.who observed the sensitivity to be only 58% after 24 h, increasing to95% after 72.
Oneof themain issueswith chromIDPawas the interpretation of thecolony colourings, which were very variable. The spectrum of coloursthat was considered a positive result was expanded in this study toinclude pink-brown as at 37 °C this increased the sensitivity (11% in-crease at 24 h of incubation) without greatly compromising specificity(3% decrease at 24 h of incubation). Interpretation of colony colourationneeds to be re-considered in future studies to ensure that maximumsensitivity is achieved. At the higher incubation temperature of 42 °Cmore P. aeruginosa strains grew producing a colour considered positive.Although the change was only slight in most cases, from a brown to apink-brown colour, this led to an increase in the sensitivity of themedi-um (98% after 24 and 72 h). A decrease in the specificity and PPV of themediumwas also observed, however, as more non-P. aeruginosa strainswere recorded as false positives. Overall, the results for this mediumwere more reliable when an incubation temperature of 37 °C wasapplied.
Media containing acetamide as the positive selective agent (Z-agar/Z-broth) have been reported to have specificity comparable to thatof nitrofurantoin broth (Szita et al., 1998), although their sensitivityhas not been evaluated. The modification of Z-agar to include addition-al nutrient supplementation permitted the growth of all of theP. aeruginosa strains, including one auxotrophic CF strain. Thismodifica-tion, however, also decreased the selectivity of the agar resulting inmZ-agar having the lowest specificity in the study. After 72 h the spec-ificity of the unmodified Z-agar was also relatively low and growth ofP. aeruginosa and non-P. aeruginosa strainswasweak on both acetamideagars making interpretation of results very difficult. In addition, as theZ-agars have to be made up in-house they would perhaps not be avery attractive choice for use in clinical or industrial settings.
4.1. Conclusions
In this study chromID Pa outperformed the other agars in terms ofspecificity but incubation time, temperature and interpretation of colo-ny colour need to be further considered in order to increase sensitivity.Chromogenic media are expensive, but if chromID Pa affords presump-tive identification of P. aeruginosa, the costs of further biochemical testsmay be off-set, as previously suggested (Laine et al., 2009). PCN and PIAare currently widely used in general selective media and have highsensitivity, but may permit the growth of other species commonly
No. of strains producing pigmenta (%)
24 h 72 h
53/58 (91) 56/58 (97)41/58 (71) 57/58 (98)47/58 (81) 58/58 (100)
Table 4Growth of non-Pseudomonas aeruginosa strains on selective media at 37 °C.
Species Source No. ofstrains
No. of strains exhibiting growth
PCNa PIAb chromID Pac Z-agar mZ-agard
24 h 72 h 24 h 72 h 24 h 72 h 24 h 72 h 24 h 72 h
Achromobacter xylosoxidans CLIN 2 2 2 2 2 0 0 1 (1 +/−) 2 2 (1 +/−) 2Acinetobacter species IND 1 0 0 0 0 0 0 1 (1 +/−) 1 1 (1 +/−) 1Bacillus species IND 1 0 0 0 0 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Burkholderia ambifaria CLIN 1 1 1 1 1 0 0 0 0 0 1 (1 +/−)
ENV 2 2 (1 +/−) 2 2 2 1 1 0 1 (1 +/−) 1 (1 +/−) 2 (1 +/−)Burkholderia cenocepacia CLIN 7 6 6 7 7 2 6 5 (5 +/−) 6 (6 +/−) 3 (3 +/−) 7 (6 +/−)
ENV 2 2 2 2 2 0 1 0 2 0 2 (2 +/−)IND 1 0 1 1 1 1 1 1 1 1 1
Burkholderia cepacia CLIN 1 1 1 1 1 0 0 1 1 1 1Burkholderia cepacia complex IND 2 1 2 (1+/−) 1 1 1 1 2 (1 +/−) 2 2 2Burkholderia contaminans CLIN 1 1 (1 +/−) 1 1 1 0 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Burkholderia dolosa CLIN 1 1 1 1 1 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Burkholderia gladioli CLIN 1 0 0 0 0 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Burkholderia lata ENV 1 1 1 1 1 1 1 1 1 1 1Burkholderia multivorans CLIN 4 4 4 4 4 2 2 3 (3 +/−) 3 (3 +/−) 3 (3 +/−) 4 (4 +/−)
ENV 1 1 1 1 1 1 1 0 0 1 (1 +/−) 1 (1 +/−)Burkholderia phymatum ENV 1 0 0 0 0 0 0 0 0 0 1 (1 +/−)Burkholderia phytofirmans ENV 1 0 0 0 0 0 0 0 0 0 1 (1 +/−)Burkholderia pyrrocinia CLIN 1 1 1 1 1 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Burkholderia stabilis CLIN 1 1 (1 +/−) 1 1 1 1 1 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Burkholderia vietnamiensis CLIN 1 1 1 1 1 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)
ENV 1 1 (1 +/−) 1 1 1 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)IND 1 1 (1 +/−) 1 1 1 0 0 1 (1 +/−) 1 1 (1 +/−) 1
Escherichia coli CLIN 1 0 0 0 0 0 0 0 0 1 (1 +/−) 1 (1 +/−)Halomonas species IND 1 0 0 0 1 0 0 0 1 (1 +/−) 1 (1 +/−) 1Ochrobactrum anthropi IND 1 0 0 0 0 1 1 1 (1 +/−) 1 1 (1 +/−) 1Pandoraea apista CLIN 2 2 2 2 2 0 0 2 (2 +/−) 2 (1 +/−) 2 (2 +/−) 2 (1 +/−)Pandoraea pnomenusa CLIN 3 3 (3 +/−) 3 (3 +/−) 3 3 0 0 3 (3 +/−) 3 3 (2 +/−) 3Pandoraea pulmonicola CLIN 2 2 2 2 2 0 0 2 (2 +/−) 2 (1 +/−) 2 (2 +/−) 2 (1 +/−)Pandoraea sputorum CLIN 4 3 3 4 4 0 0 4 (4 +/−) 4 (1 +/−) 4 (4 +/−) 4 (1 +/−)Pantoea agglomerans IND 1 0 0 0 0 0 0 0 0 1 (1 +/−) 1 (1 +/−)Pseudomonas alcaligenes ENV 1 1 1 0 1 0 0 0 0 1 (1 +/−) 1 (1 +/−)Pseudomonas aureofaciens ENV 1 1 1 1 1 1 1 0 0 1 (1 +/−) 1 (1 +/−)Pseudomonas fluorescens ENV 1 0 0 1 (1 +/−) 1 (1+/−) 0 0 0 0 0 0
IND 2 2 2 2 2 0 2 0 0 0 0Pseudomonas mendocina ENV 1 1 1 1 (1 +/−) 1 1 1 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Pseudomonas oleovorans IND 1 1 (1 +/−) 1 0 0 0 0 1 (1 +/−) 1 1 (1 +/−) 1Pseudomonas pseudoalcaligenes IND 1 1 (1 +/−) 1 0 1 0 0 0 1 (1 +/−) 1 (1 +/−) 1Pseudomonas putida ENV 1 1 (1 +/−) 1 1 1 0 0 0 0 0 0
IND 7 6 (1 +/−) 7 (1 +/−) 7 7 3 4 6 (6 +/−) 6 (3 +/−) 7 (7 +/−) 7 (4 +/−)Pseudomonas species IND 8 7 (1 +/−) 8 (2 +/−) 6 7 2 3 1 (1 +/−) 5 (5 +/−) 6 (6 +/−) 7 (6 +/−)Ralstonia insidiosa CLIN 1 0 0 0 0 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Ralstonia mannitolilytica CLIN 2 0 0 0 0 0 0 2 (2 +/−) 2 (2 +/−) 2 (2 +/−) 2 (2 +/−)Ralstonia pickettii CLIN 3 0 0 0 0 0 0 3 (3 +/−) 3 (3 +/−) 3 (3 +/−) 3 (1 +/−)Serraria marcescens ENV 2 0 0 2 2 0 0 − − − −
IND 1 0 0 1 1 0 0 – – – –
NA 1 0 0 1 1 0 0 – – – –
Staphylococcus aureus CLIN 2 0 0 0 0 0 0 0 0 1 (1 +/−) 2 (2 +/−)Staphylococcus epidermidis CLIN 1 0 0 0 0 0 0 0 0 1 (1 +/−) 1 (1 +/−)Stenotrophomonas maltophilia CLIN 1 0 0 0 1 0 0 0 1 (1 +/−) 1 (1 +/−) 1 (1 +/−)Stenotrophomonas species IND 1 0 0 1 1 0 0 0 1 (1 +/−) 1 (1 +/−) 1Total 59/90
(13 +/−)e63/90(7 +/−)
65/90(2 +/−)
69/90(1 +/−)
18/90 27/90 50/86(46 +/−)
64/86(39 +/−)
68/86(61 +/−)
81/86(51 +/−)
Specificity (%) 34 30 28 23 80 70 42 28 26 6
CLIN, clinical; ENV, environmental; IND, industrial; NA, source unknown.a PCN, Pseudomonas CN selective agar.b PIA, Pseudomonas isolation agar.c chromID Pa, chromID® P. aeruginosa; a positive result on chromID Pa was taken as growth accompanied by a red/pink/pink-brown/purple colouration.d mZ-agar, modified Z-agar.e Weakly growing strains (+/−) were recorded as an overall positive and are indicated in the table.
13R. Weiser et al. / Journal of Microbiological Methods 99 (2014) 8–14
encountered in CF and from industrial settings. Even though increasingthe incubation temperature to 42 °C may improve specificity andPPV values, further biochemical tests and/or genotypic methods are re-quired for accurate species identification when using these media. Z-agar had lower specificity than previously reported (Szita et al., 2007;Szita et al., 1998) and is likely to be unsuitable for application in clinicalor industrial environments without further modification.
Supplementary data to this article can be foundonline at http://dx.doi.org/10.1016/j.mimet.2014.01.010.
Acknowledgements
Rebecca Weiser acknowledges funding from the Biotechnology andBiological Sciences Research Council PhD funding (Doctoral TrainingGrant BB/F016557/1) with CASE sponsorship from the Unilever Re-search andDevelopment, Port Sunlight,Wirral, England, UK. EuroCareCFis acknowledged for the EuroCareCF strain panel (highlighted in Supple-mentary material Table S1): European Coordination Action for Researchin Cystic Fibrosis; EC FP6 project no. LSHM-CT-2005-018932.
14 R. Weiser et al. / Journal of Microbiological Methods 99 (2014) 8–14
References
Bador, J., Amoureux, L., Duez, J.-M., Drabowicz, A., Siebor, E., Llanes, C., Neuwirth, C., 2011.First description of a RND-typemultidrug efflux pump in Achromobacter xylosoxidans:AxyABM. Antimicrob. Agents Chemother. 55, 4912–4914.
Deschaght, P., Van daele, S., De Baets, F., Vaneechoutte, M., 2011. PCR and the detection ofPseudomonas aeruginosa in respiratory samples of cystic fibrosis patients. A literaturereview. J. Cyst. Fibros. 10, 293–297.
De Vos, D., Lim, A., Pirnay, J.P., Struelens, M., Vandenvelde, C., Duinslaeger, L., Vanderkelen,A., et al., 1997. Direct detection and identification of Pseudomonas aeruginosa in clin-ical samples such as skin biopsy specimens and expectorations by multiplex PCRbased on two outer membrane lipoprotein genes, oprI and oprL. J. Clin. Microbiol.35, 1295–1299.
FDA, 1998. Bacteriological analytical manual, 8th edition. (Revision A, Chapter 23 [On-line]. Available at: http://www.fda.gov/food/scienceresearch/laboratorymethods/bacteriologicalanalyticalmanualbam/ucm073598.htm [Accessed: 28/06/12]).
Gomez, M., Prince, A., 2007. Opportunistic infections in lung disease: Pseudomonasinfection in cystic fibrosis. Curr. Opin. Pharmacol. 7, 244–251.
Hauser, A.R., Jain, M., Bar-Meir, M., McColley, S.A., 2011. Clinical significance of microbialinfection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 24, 29–70.
Hogardt, M., Ulrich, J., Riehn-Kopp, H., Tümmler, B., 2009. EuroCareCF quality assessmentof diagnostic microbiology of cystic fibrosis isolates. J. Clin. Microbiol. 47, 3435–3438.
Jimenez, L., 2007. Microbial diversity in pharmaceutical product recalls andenvironments. PDA J. Pharm. Sci. Technol. 61, 383–399.
Jimenez, L., Ignar, R., Smalls, S., Grech, P., Hamilton, J., Bosko, Y., English, D., 1999. Molec-ular detection of bacterial indicators in cosmetic/pharmaceuticals and raw materials.J. Ind. Microbiol. Biotechnol. 22, 93–95.
Jimenez, L., Smalls, S., Ignar, R., 2000. Use of PCR analysis for detecting low levels of bacteriaandmold contamination in pharmaceutical samples. J. Microbiol. Methods 41, 259–265.
Kerr, K.G., Snelling, A.M., 2009. Pseudomonas aeruginosa: a formidable and ever-presentadversary. J. Hosp. Infect. 72, 338–344.
King, E.O., Ward, M.K., Raney, D.E., 1954. Two simple media for the demonstration ofpyocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307.
Kodaka, H., Iwata, M., Yumoto, S., Kashitani, F., 2003. Evaluation of a new agarmedium containing cetrimide, kanamycin and nalidixic acid for isolation andenhancement of pigment production of Pseudomonas aeruginosa in clinicalsamples. J. Basic Microbiol. 43, 407–413.
Laine, L., Perry, J.D., Lee, J., Oliver, M., James, A.L., De La Foata, C., Halimi, D., et al.,2009. A novel chromogenic medium for isolation of Pseudomonas aeruginosafrom the sputa of cystic fibrosis patients. J. Cyst. Fibros. 8, 143–149.
Lalkhen, A.G., McCluskey, A., 2008. Clinical tests: sensitivity and specificity. Contin. Educ.Anaesth., Crit. Care Pain 8, 221–223.
Leisinger, T., Margraff, R., 1979. Secondary metabolites of the fluorescent pseudomonads.Microbiol. Rev. 43, 422–442.
Li, X.-Z., Nikaido, H., 2009. Efflux-mediated drug resistance in bacteria: an update.Drugs 69, 1555–1623.
Lilly, H.A., Lowbury, E.J., 1972. Cetrimide-nalidixic acid agar as a selective medium forPseudomonas aeruginosa. J. Med. Microbiol. 5, 151–153.
Morrison, A.J., Wenzel, R.P., 1984. Epidemiology of infections due to Pseudomonasaeruginosa. Rev. Infect. Dis. 6, S627–S642.
Perry, J.D., Freydière, A.M., 2007. The application of chromogenic media in clinicalmicrobiology. J. Appl. Microbiol. 103, 2046–2055.
Reyes, E.A., Bale, M.J., Cannon, W.H., Matsen, J.M., 1981. Identification of Pseudomonasaeruginosa by pyocyanin production on Tech agar. J. Clin. Microbiol. 13, 456–458.
Rose, H., Baldwin, A., Dowson, C.G., Mahenthiralingam, E., 2009. Biocidesusceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother.63, 502–510.
Smirnov, V.V., Kiprianova, E.A., 1990. Bacteria of the Pseudomonas genus. NaukovaDumka, Kiev, USSR 100–111.
Spilker, T., Coenye, T., Vandamme, P., LiPuma, J.J., 2004. PCR-based assay fordifferentiation of Pseudomonas aeruginosa from other Pseudomonas species recoveredfrom cystic fibrosis patients. J. Clin. Microbiol. 42, 2074–2079.
Stover, C.K., Pham, X.Q., Erwin, A.L., Mizoguchi, S.D., Warrener, P., Hickey, M.J.,Brinkman, F.S.L., et al., 2000. Complete genome sequence of Pseudomonasaeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964.
Sutton, S., Jimenez, L., 2012. A review of reported recalls involving microbiologicalcontrol 2004–2011 with emphasis on FDA considerations of ‘objectionable or-ganisms’. Am. Pharm. Rev. 15, 42–57.
Szita, G., Tabajdi, V., Fábián, A., Biró, G., Reichart, O., Körmöczy, P.S., 1998. A novel,selective synthetic acetamide containing culture medium for isolating Pseudomonasaeruginosa from milk. Int. J. Food Microbiol. 43, 123–127.
Szita, G., Gyenes, M., Soós, L., Rétfalvi, T., Békési, L., Csikó, G., Bernáth, S., 2007. Detection ofPseudomonas aeruginosa in water samples using a novel synthetic medium andimpedimetric technology. Lett. Appl. Microbiol. 45, 42–46.
Thomas, S.R., Ray, A., Hodson, M.E., Pitt, T.L., 2000. Increased sputum amino acidconcentrations and auxotrophy of Pseudomonas aeruginosa in severe cysticfibrosis lung disease. Thorax 55, 795–797.
Winstanley, C., Langille, M.G.I., Fothergill, J.L., Kukavica-Ibrulj, I., Paradis-Bleau, C.,Sanschagrin, F., Thomson, N.R., et al., 2009. Newly introduced genomicprophage islands are critical determinants of in vivo competitiveness in theLiverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 19, 12–23.