E Wild -type of 35S:MYB106-SRDX F flowers and torenia leaf. · PDF fileSupplemental Data....
Transcript of E Wild -type of 35S:MYB106-SRDX F flowers and torenia leaf. · PDF fileSupplemental Data....

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
1 / 18
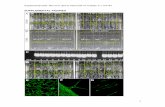
Supplemental Figure 1. Cuticle permeability
of 35S:MYB106-SRDX Arabidopsis flowers
and torenia leaf.
(A) Wild type stained with Toluidine Blue (TB).
No staining was observed.
(B) Weak-phenotype line of
35S:MYB106-SRDX stained with TB. The tops
and margins of sepals were stained; the anther
was also stained.
(C) and (D) Siliques grew on wild-type plants
but did not grow on 35S:MYB106-SRDX.
(E) and (F) Wild-type torenia (E) and 35S:MYB106-SRDX torenia (F), stained with TB.
Bars indicate 1 mm in (A) and (B) and 2 mm in
(E) and (F).
Wild-type 35S:MYB106-SRDX
A B
Wild-type 35S:MYB106-SRDX (mild)
C
D
35S:MYB106-
SRDX (torenia)
Wild-type
(torenia)E F

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
2 / 18
A
B

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
3 / 18
Supplemental Figure 2. Expression and motif analyses based on public
microarray data or conserved motifs among MYB subgroup 9 genes/proteins,
respectively.
(A) Comparison of microarray data of MYB106, MYB16 and MYB17 along with
developmental stages using AtGenExpress Visualization Tool
(http://jsp.weigelworld.org/expviz/expviz.jsp). X and Y axes represent different
developmental stages or signal intensity, respectively.
(B) One-by-one comparison of microarray data of MYB106, MYB16 and MYB17
along with developmental stages using ATTED-II database (Obayashi et al., 2009).
Top-left, top-right and bottom panels represent comparison of MYB16 (X axis) and
MYB17 (Y axis), MYB106 (X axis) and MYB17 (Y axis), and MYB16 (X axis) and
MYB106 (Y axis), respectively. Each axis represents signal intensity.
(C) Comparison of conserved protein motifs among MYB subgroup 9 proteins
collected from various species. The analysis was performed by SALAD database
(Mihara et al., 2008). At, Arabidopsis thaliana; ARALYDRAFT, Arabidopsis lyrata; Gt,
Gentiana triflora; VIT, Vitis vinifera; MTR, Medicago truncatula; Am, Antirrhinum
majus; Cs, Cucumis sativus; Pt, Populus trichocarpa; Gm, Glycine max; Dac,
Daucus carota; Tf, Torenia fournieri; Pf, Perilla frutescens; RCOM, Ricinus
communis; La, Lupinus albus; Md, Malus x domestica; Rh, Rosa hybrid cultivar; If,
C M YB 106
M YB 16
M YB 17
M YB 106 /
M YB 16
group
M YB 17
group
Md_MYB13
At_MYB106
Am_M IXTA
Pt_MYB138
Pt_MYB038
Am_MYBML1
Am_MYBML3
Hi_M IXTA1
Am_MYBML2
Ah_MYB25
Dac _MYB2
At_MYB16
Pt_MYB083
Dec _MYBML1
Ph_MYB1
Zm_MYB34
Md_MYB14
At_MYB17
Tf_MYBML3
Pt_MYB187
Pt_MYB077
C

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
4 / 18
Iris fulva; Zm, Zea mays; SORBIDRAFT, Sorghum bicolor; Os, Oryza sativa
Japonica Group; PHYPADRAFT, Physcomitrella patens; Bd, Brachypodium
distachyon; Ta, Triticum aestivum; Hv, Hordeum vulgare; Dec, Dendrobium
crumenatum; Gh, Gossypium hirsutum; Hi, Humulus lupulus; Ph, Petunia x hybrid;
AmMIXTA, CAA55725; Am-MYBML1, CAB43399; Am-MYBML2, AAV70655;
Ph-MYB1, CAA78386; Pt-MYB083, XP_002323871; Dc-MYB2, BAF49441;
Pt-MYB187, POPTRDRAFT_572225; Md-MYB14, AAZ20433; Pt-MYB077,
POPTRDRAFT_575814; Zm-MYB34, AAL90652; Md-MYB13, AAZ20432;
Dec-MYBML1, ADD64500; Gh-MYB25, ACJ07153; Pt-MYB038,
POPTRDRAFT_567011; Pt-MYB138, POPTRDRAFT_564238; Hi-MIXTA1,
BAF46264.

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
5 / 18
Supplemental Figure 3. Phenotypes
caused by constitutive expression of
MYB16-SRDX.
(A) Fused leaves of 4-week-old
35S:MYB16-SRDX plant.
(B) Scanning electron micrographs of fused
flower of 35S:MYB16-SRDX plants.
(C) Surface of stem of 35S:MYB16-SRDX
plant. Cuticular wax was reduced.
Bars indicate 5 mm in (A), 0.2 mm in (B),
33.33 μm in (C).

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
6 / 18
M
YB
16
pro
:GU
S
I
I
J
G H
MY
B1
06
pro
:GU
S A B C
D
F
E
Supplemental Figure 4. Promoter activities of MYB106 and MYB16. (A) to (E) GUS reporter activity (blue color) driven by the promoter of MYB106 in 3-week-old plants (A), trichomes (B), stem and pedicel (C), stem (D), and dehiscence zone between carpel and pedicel (E). (F) to (J) GUS reporter activity driven by the promoter of MYB16 in 3-week-old plants (F), a young rosette leaf (G), stem (H), epidermis of rosette leaf (I) and mesophyll cell (J). Bars indicate 0.1 mm in (B) to (E) and (G) to (J), 2 mm in (A) and (F).

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
7 / 18
Supplemental Figure 5.
Promoter activities of
MYB106 and MYB16 in
floral organs.
GUS reporter activity (blue color) driven by the promoter of MYB106 or MYB16 in buds and flower. Asterisks and circles indicate petals and stamens showing GUS activity, respectively. Bars indicate 0.1 mm.
MYB16pro:GUSMYB106pro:GUS
Stage 10
Stage 11
Stage 12
early
Stage 12
late
Stage 13
*
*
**
*
*
*
* *
*
*
*
*
*
○
○
○
○
○
○
○
○
○
○
○
○
○
○

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
8 / 18
35S:MYB106 -RNAi
A Seedlings Buds B
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
Rela
tive a
mo
un
t o
f M
YB
106 m
RN
A
*
**
35S:MYB106-RNAi
D
D
35S:MYB106-RNAi
0
0.2
0.4
0.6
0.8
1
1.2
1.4
Rela
tive a
mo
un
t o
f M
YB
16 m
RN
A
*
Seedlings Buds
C E
Supplemental Figure 6.
Plants with reduced levels
of MIXTA-like MYBs.
(A) and (B) Quantitative
RT-PCR analysis of 3’
non-coding region of MYB106
(A) and coding region of
MYB16 (B) in 3-week-old
plants (n = 4 to 6) and buds of
35S:MYB106RNAi and
wild-type plants (n = 4 to 6).
Expression of MYB106 and
MYB16 was reduced in the
RNAi line. Error bar represent
standard deviation. Single
asterisks indicate P < 0.05 in
Welch’s t-test. (C) Rosette leaf
and magnified view of
trichome on it of
35S:MYB106-RNAi
Arabidopsis stained with TB.
(D) Flower of
35S:MYB106-RNAi plants
stained by TB. Organ
adhesion occurs inside flower.
Edge of sepal and petal is
stained by TB. (E) Entire
35S:MYB106-RNAi plants,
which are almost sterile due to
organ adhesion inside the
flower. Bars indicate 100μm in
(C), 1mm in (D).

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
9 / 18
Supplemental Figure 7.
Constitutive expression of
SHN3-SRDX caused reduction of
cuticular wax with severe
morphological changes.
(A) Fused leaves of 3-week-old
35S:SHN3-SRDX Arabidopsis.
(B) Fused leaves and buds of
10-week-old 35S:SHN3-SRDX plant.
(C) to (E) Surface of stem (C) and
petal (E) of 35S:SHN3-SRDX plant
observed by SEM. Fewer wax
crystals and cutin nanoridges than
wild type (D) were observed.
(F) TB uptake per gram protein.
Error bars represent standard error
(n=8). Double asterisks represent
P<0.01 by Welch’s t-test. (G) Leaf of
35S:WIN1-SRDX torenia stained
with TB. Bars indicate 5 mm in (A)
and (B) and 2 mm in (G) 33.33 μm in
(C), 10 μm in (D) and (E).
35S:SHN3-SRDXA
B
35S:SHN3-SRDX
(Petal)
Wild-type
(Petal)D
E
35S:SHN3-SRDX
C 35S:SHN3-SRDX(Stem)
10ul
0.00
0.02
0.04
0.06
0.08
0.10
TB
/pro
tein
(g/g
) **F G 35S:WIN1-SRDX
(torenia)

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
10 / 18
Supplemental Table 1. Frequency of observed phenotypes for four transgenes
Severe Intermediate Mild Wild-type like Total
35S:MYB106-SRDX 7 7 16 1 31
35S:WIN1-SRDX 17 34 1 13 65
MYB106pro:MYB106-SRDX 0 9 7 12 28
35S:MYB106-RNAi 0 14 24 8 46
Severe, intermediate, mild phenotypes represent organ adhesions in whole plant, immature
trichome and organ adhesion in limited tissues, over-branched trichome, respectively. “Total”
represents the number of plants examined for each transgene.

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
11 / 18
Supplemental Table 2. Branch numbers of leaf trichomes
Branch number of trichome Total*
1 2 3 4 5 6 7 8
Wild-type 0.60 7.23 90.06 2.11 0.00 0.00 0.00 0.00 664
35S:MYB106-SRDX 5.99 22.12 53.92 13.36 4.61 0.00 0.00 0.00 217
MYB106pro:MYB106-SRDX 98.59 1.41 0.00 0.00 0.00 0.00 0.00 0.00 142
myb106-1 0.47 10.54 59.95 23.65 5.39 0.00 0.00 0.00 427
myb106-2 2.03 18.06 38.60 23.02 17.38 0.90 0.00 0.00 443
MYB16amiRNA 0.25 2.78 81.27 15.44 0.25 0.00 0.00 0.00 395
MYB16amiRNA myb106-2 0.23 0.45 9.01 15.09 50.00 20.50 4.50 0.45 444
Numbers in table are percentage of trichomes with indicated branch number.
*Total number of trichomes on leaf 3 and 4 of five plants.

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
12 / 18
Supplemental Table 3. Down regulated gene groups in both MYB106-SRDX and
WIN1-SRDX plants
GO term Number
of
genes a
Downregulated
genes b
P-value c Odds
ratio d
Cuticle development 45 15 3.63E-11 10.5386
Wax biosynthetic process 16 3 0.01482 5.92795
Cutin biosynthetic process 10 5 2.01E-05 15.8079
Very-long-chain fatty acid
metabolic process
43 16 1.62E-12 11.764
Fatty acid elongase activity 6 4 4.62E-05 21.0772
Carboxylesterase activity 129 14 8.86E-05 3.43117
Lipid metabolic process 246 21 5.81E-05 2.69891
Transferase activity,
transferring acyl groups other
than amino-acyl groups
73 10 0.0001437 4.33092
Lipid transport 157 13 0.001836 2.61787
Hydrolase activity, acting on
ester bonds
172 13 0.003944 2.38956
Secondary cell wall biogenesis 35 6 0.0009878 5.41984
Plant-type cell wall
modification involved in
multidimensional cell growth
25 3 0.04602 3.79389
Response to auxin stimulus 262 34 3.91E-08 2.96944

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
13 / 18
a Number of genes included in each GO category
b Number of genes downregulated in both 35S:MYB106-SRDX and 35S:WIN1-SRDX
plants c P-value of Fisher’s exact test
d Ratio between actually observed number and expected number by chance

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
14 / 18
Supplemental Table 4. Changes in expression of cuticle related genes by microarray analysis.Locus Common name Short description #1 #2 #3 #4AT4G34520 KCS18, FAE1 3-ketoacyl-CoA synthase 18 1.01 0.95 0.83 0.73
AT1G62340 ALE1 PA-domain containing subtilase family protein 1 0.95 0.94 0.68
AT1G49430 LRD2, LACS2 long-chain acyl-CoA synthetase 2 0.37 0.2 3.04 1.96
AT1G01610 ATGPAT4, GPAT4 glycerol-3-phosphate acyltransferase 4 0.35 0.12 3.31 1.46
AT1G36160 GSD1, PAS3, GK, EMB22, AT-ACC1, ACC1 acetyl-CoA carboxylase 1 0.62 0.42 1.2 1.45
AT1G68530 KCS6, CER6, POP1, G2, CUT1 3-ketoacyl-CoA synthase 6 0.62 0.31 1.55 1.88
AT1G02205 CER1 Fatty acid desaturase 0.24 0.08 2.79 3.33
AT3G10570 CYP77A6 cytochrome P450, family 77, subfamily A, polypeptide 6 0.39 0.45 10.9 14.63
AT1G01600 CYP86A4 cytochrome P450, family 86, subfamily A, polypeptide 4 0.37 0.18 9.34 7.72
AT2G04570 GDSL-like Lipase/Acylhydrolase superfamily protein 0.53 0.13 20.03 13.25
AT1G63710 CYP86A7 cytochrome P450, family 86, subfamily A, polypeptide 7 0.63 0.11 4.11 10.13
AT2G38110 ATGPAT6, GPAT6 glycerol-3-phosphate acyltransferase 6 0.21 0.18 2.87 5.52
AT5G23940 PEL3, DCR, EMB3009 HXXXD-type acyl-transferase family protein 0.44 0.64 1.38 1.87
AT5G10480 PEP, PAS2 Protein-tyrosine phosphatase-like, PTPLA 0.5 0.7 1.17 2.36
AT1G67730 ATKCR1, YBR159, KCR1 beta-ketoacyl reductase 1 0.45 0.84 1.02 1.43
AT4G33790 G7, FAR3, CER4 Jojoba acyl CoA reductase-related male sterility protein 0.04 0.44 1.32 4.67
AT1G10670 ACLA-1 ATP-citrate lyase A-1 0.63 1 1.08 0.95
AT4G34250 KCS16 3-ketoacyl-CoA synthase 16 0.12 0.1 0.12 0.95
AT2G16280 KCS9 3-ketoacyl-CoA synthase 9 0.26 0.42 0.45 0.61
AT2G15090 KCS8 3-ketoacyl-CoA synthase 8 0.09 0.23 0.25 1.62
AT4G28110 ATMYB41 myb domain protein 41 0.21 0.55 0.23 1.74
AT5G23190 CYP86B1 cytochrome P450, family 86, subfamily B, polypeptide 1 0.06 0.39 0.07 0.61
AT5G58860 CYP86A1 cytochrome P450, family 86, subfamily A, polypeptide 1 0.04 0.21 0.05 1.08
AT4G24510 VC2, VC-2, CER2 HXXXD-type acyl-transferase family protein 0.21 0.01 2.66 0.75
AT2G20300 ALE2 Protein kinase superfamily protein 0.71 0.66 1.07 0.8
AT1G25450 KCS5, CER60 3-ketoacyl-CoA synthase 5 0.18 0.16 1.31 1.02
AT1G01120 KCS1 3-ketoacyl-CoA synthase 1 0.13 0.08 2.75 0.66
AT4G00360 ATT1, CYP86A2 cytochrome P450, family 86, subfamily A, polypeptide 2 0.21 0.25 2.08 0.86
AT2G47240 CER8, LACS1 AMP-dependent synthetase and ligase family protein 0.32 0.35 0.68 2
AT1G51500 AtABCG12, WBC12, ABCG12, D3, CER5, ABC-2 type transporter family protein 0.47 0.2 0.74 1.63
AT2G34770 ATFAH1, FAH1 fatty acid hydroxylase 1 0.2 0.35 0.64 0.95
AT4G00400 AtGPAT8, GPAT8 glycerol-3-phosphate acyltransferase 8 0.05 0.1 0.99 0.73
AT2G45970 CYP86A8, LCR cytochrome P450, family 86, subfamily A, polypeptide 8 0.18 0.31 0.82 0.9
AT2G26250 KCS10, FDH 3-ketoacyl-CoA synthase 10 0.18 0.21 1.13 2.06
AT4G24140 BDG3 alpha/beta-Hydrolases superfamily protein 0.14 0.27 0.91 1.41
AT3G55360 ECR, CER10, ATTSC13, TSC13 3-oxo-5-alpha-steroid 4-dehydrogenase family protein 0.57 0.66 1.06 1.11
AT5G49070 KCS21 3-ketoacyl-CoA synthase 21 0.86 1.09 1.25 0.58
AT1G19440 KCS4 3-ketoacyl-CoA synthase 4 1.01 0.77 1.4 0.88
AT5G57800 CER3, FLP1, YRE, WAX2 Fatty acid hydroxylase superfamily 0.52 0.4 1.89 0.43
AT2G27690 CYP94C1 cytochrome P450, family 94, subfamily C, polypeptide 1 0.43 0.08 3.14 0.2
AT3G28910 AtMYB30, MYB30 myb domain protein 30 0.67 0.66 1.68 0.71
AT5G47120 ATBI-1, ATBI1 BAX inhibitor 1 1.22 0.97 1.18 1.35
AT3G54010 DEI1, PAS1 FKBP-type peptidyl-prolyl cis-trans isomerase family protein 1.02 1.11 1.2 1.27
AT1G71160 KCS7 3-ketoacyl-CoA synthase 7 2.62 1.85 1.03 2.87
AT1G69500 CYP704B1 cytochrome P450, family 704, subfamily B, polypeptide 1 1.25 4.46 1.43 2.12
AT1G57750 MAH1, CYP96A15 cytochrome P450, family 96, subfamily A, polypeptide 15 1.38 2.42 1.39 0.74
AT3G60500 G3, CER7 3'-5'-exoribonuclease family protein 1.47 1.54 1.12 0.9
AT2G33510 AtCFL1 unknown protein 1.51 1.81 0.68 0.94
AT4G34510 KCS17, KCS2 3-ketoacyl-CoA synthase 17 0.95 1.39 0.55 1.11
AT4G14070 AAE15 acyl-activating enzyme 15 1.34 1.06 0.64 1.1
AT3G12990 RRP45A ribonuclease PH45A 1.11 1.08 0.76 1.24
AT5G37300 WSD1 O-acyltransferase (WSD1-like) family protein 0.44 0.81 0.29 8.48
AT2G46720 KCS13, HIC 3-ketoacyl-CoA synthase 13 1.22 0.32 0.72 1.17
AT3G61150 HD-GL2-1, HDG1 homeodomain GLABROUS 1 1.46 0.56 0.69 1.5
#1 35S_MYB106SRDX_3wk_seedling/Col0_3wk_seedling
#2 35S_WIN1SRDX_3wk_seedling/Col0_3wk_seedling
#3 WIN1_ox_2wk_seedling/vector_control_2wk_seedling
#4 MYB106VP16_seedling2/Col0_seedling_Oshima
Cuticle related genes were collected by the Gene ontology terms as cuticle development, cutin biosynthetic process, wax biosynthetic process, alkane 1-
monooxygenase activity, very long-chain fatty acid biosynthetic process, fatty acid elongation, fatty acid elongation, very long-chain fatty acid
biosynthetic process and very long-chain fatty acid metabolic process besides AT2G45970 (Wellesen et al. 2001), AT2G04570 (Kannangara et al. 2007),
AT5G10480 (Bach et al. 2008), AT3G61150, AT2G33510(Wu et al. 2011), AT4G24140 (Kurdyukov et ol. 2006, Shi et al. 2011).

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
15 / 18
Supplemental table 5. Phenotypes of loss-of-function plants
trichome petal cell outgrowth cuticle deficiency
nok-122 over-branching Folkers et al. 1997
myb106-1 over-branching,
immature
Guildinf and Marks 2010
Jakoby et al. 2008
petal and filament
nanoridge this study
myb106-2
over-branching,
less-branching,
immature
this study petal and filament
nanoridge this study
MYB16amiRNA over-branching this study filament nanoridge this study
MYB16amiRNA myb106-2 over-branching and
immature this study supressed this study
petal and filament
nanoridge this study
35S:MYB106-SRDX
over-branching,
less-branching,
immature
this study supressed this study stem, leaf, petal
nanoridge, trichome this study
35S:MYB16-SRDX stem this study
MYB106pro:MYB106-SRDX less-branching,
immature this study supressed this study
stem, petal, stamen,
carpel, trichome this study
Antirrhinum mixta supressed Noda et al. 1994
Petunia phmyb1 supressed Baumann et al. 2007
Supplemental table 6. Phenotypes of gain-of-function plants and references
trichome petal cell outgrowth ectopic formation of specific cuticle
MYB106-VP16 less-branching,
early maturation this study nanoridge on leaves this study
MYB106ox less-branching,
early maturation this study nanoridge on carpel this study
MYB16ox enhanced in tobacco Baumann et al. 2007
AmMYBML1ox ectopic formation in
tobacco
Rerez-Rodriguez et al.
2004
glandular trichome
formation in tobacco
Rerez-Rodriguez et al.
2004
AmMYBML2ox enhanced in tobacco Baumann et al. 2007
PhMYB1ox enhanced in tobacco Baumann et al. 2007
MIXTAox trichome formation in
tobacco Glover et al. 1998

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
16 / 18
Supplemental Table 7. Primers used in this study.
Name Sequenses (5' - 3') Purpose
AT3G01140N gATGGGCAGATCGCCATGTTGTGATAAGGC 35S:MYB106SRDX, 35S:MYB106, 35S:MYB106VP16, MYB106pro:MYB106SRDX
AT3G01140C GAACATCGTCGCGGAATCGGACGGTGAAGA 35S:MYB106SRDX, 35S:MYB106VP16, MYB106pro:MYB106SRDX
AT1G15360F ATGGTACAGACGAAGAAGTTCAG 35S:WIN1SRDX, 35S:WIN1
AT1G15360R GTTTGTATTGAGAAGCTCCTCTATC 35S:WIN1SRDX
At5G25390F ATGGTACATTCGAAGAAGTTCCG 35S:SHN3SRDX
At5G25390R GACCTGTGCAATGGATCCAG 35S:SHN3SRDX
AT5G15310N gATGGGTAGATCACCGTGTTGTGACAAATT 35S:MYB16SRDX, MYB16pro:MYB16SRDX
AT5G15310C GAACATCGGTGAATCCGACGGTGAAGGATC 35S:MYB16SRDX, MYB16pro:MYB16SRDX
AT1G15360.1SC TTAGTTTGTATTGAGAAGCTCCTCTATCAT 35S:WIN1
AT3G01140.1SC TCAGAACATCGTCGCGGAATCGGACGGTGA 35S:MYB106
AT5G15310pFA GGCGCGCCAATTTGAGACACATAAACATCTAAGCA MYB16pro:MYB16SRDX
AT5G15310pRB CGCGGATCCTGTTTTGAGAGCAAAGAAATAAGA MYB16pro:MYB16SRDX
AT5G15310pF3000 GGGGACAACTTTGTATAGAAAAGTTGAATTTGAGACACATAAACATCTAAGCA MYB16pro:GUS, MYB16pro:LUC
AT5G15310pR GGGGACTGCTTTTTTGTACAAACTTGGCATTGTTTTGAGAGCAAAGAAATAAGA MYB16pro:GUS, MYB16pro:LUC
AT3G01140pFA GGCGCGCCTAATACTAAATTTATAAATGATGGAAA MYB106pro:MYB16SRDX
AT3G01140pRB CGCGGATCCTCAATTGGATGGGGTCGAGAGATA MYB106pro:MYB16SRDX
AT3G01140pF3000 GGGGACAACTTTGTATAGAAAAGTTGTAATACTAAATTTATAAATGATGGAAA MYB106pro:GUS, MYB106pro:LUC
AT3G01140pR2 GGGGACTGCTTTTTTGTACAAACTTGGCATTGTTTTCCCACACAAGTTATTAG MYB106pro:GUS, MYB106pro:LUC
AT1G15360pF3000 GGGGACAACTTTGTATAGAAAAGTTGTCTGAAGTCTTGTTTCTTCATTATTCT WIN1pro:GUS, WIN1pro:LUC
AT1G15360pR GGGGACTGCTTTTTTGTACAAACTTGGCATTCTTACTTACTCTGTGATGTTTGT WIN1pro:GUS, WIN1pro:LUC
AT1G01600pF2000 GGGGACAACTTTGTATAGAAAAGTTGCTTGTGTGTGGTCCGAAGAAGATGAGA CYP86A4pro:LUC
AT1G01600pR GGGGACTGCTTTTTTGTACAAACTTGGCATGTAGCTCTTTTATTATTGTTTCCC CYP86A4pro:LUC
Underlines indicate attB sequences. Double underlines indicate restriction enzyme recognition site.

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
17 / 18
Supplemental Table 8. Primer sequences for RT-PCR.
Name Sequences (5' - 3') Purpose
FDHf TGCTGACGACCGTAGCTTCA quantitative RT-PCR for FDH
FDHr TTGTCTTGAGAGCTTCACCTCCAA quantitative RT-PCR for FDH
LCRf TGGTAACGGCGTTTCAAGTTAGG quantitative RT-PCR for LCR
LCRr GTTGGCCAAAGAACGGCGTATTTA quantitative RT-PCR for LCR
KCS1f GCGGTTTGGAAAGCGTTACG quantitative RT-PCR for KCS1
KCS1r CAATCGAACCAGCCCAAGCA quantitative RT-PCR for KCS1
CER2f CTTGTCTACGATCACGTTCTTGGT quantitative RT-PCR for CER2
CER2r TCCAAGAATATGGGCCCAACTCA quantitative RT-PCR for CER2
CER1f GTGAATGTCCACTCCTGTGAGA quantitative RT-PCR for CER1
CER1r AAGAAGGGATGTGCCACACTCA quantitative RT-PCR for CER1
LACS2f AAAGGCCACAGCTCCTCCAA quantitative RT-PCR for LACS2
LACS2r CCATGGACCTCTTTGCTTCTGA quantitative RT-PCR for LACS2
CYP86A4f GAAGAAGAGCGACGGTGTCT quantitative RT-PCR for CYP86A4
CYP86A4r TAAGTAAACGGCGACGCCTTCA quantitative RT-PCR for CYP86A4
CYP86A7f AGCTGGACGTGACACGTCAT quantitative RT-PCR for CYP86A7
CYP86A7r GACTCGTGGGTTAAGACTAACCA quantitative RT-PCR for CYP86A7
CYP77A6f GAGCAAGAGCTCTCGAGGTT quantitative RT-PCR for CYP77A6
CYP77A6r TCTCCGTCGTCTCTCGATGA quantitative RT-PCR for CYP77A6
2g04570f TCATCAAGAACCCTTCCTCCTTTG quantitative RT-PCR for AT2g04570
2g04570r GACAACCATATCCCATCTCGAACA quantitative RT-PCR for AT2g04570
MYB106f CAAACCAAGGAAACGGAGACCAA quantitative RT-PCR for MYB106
MYB106r CTGCTATCCGTAGGGATTCCTA quantitative RT-PCR for MYB106
MYB106-3'f ACCGTCCGATTCCGCGACGA quantitative RT-PCR for 3' noncoding region of
MYB106
MYB106-3'r GCAGGTTTGCTACATGCAATTCTGC quantitative RT-PCR for 3' noncoding region of
MYB106
WIN1/SHN1f AGCTCCTCCCAAGAGACTAC quantitative RT-PCR for WIN1/SHN1
WIN1/SHN1r TTTCAACTTCAGTGGTCGGAGCAA quantitative RT-PCR for WIN1/SHN1
MYB16f GATGGGTAGATCACCGTGTTGTGACA
AATT
quantitative RT-PCR for MYB16

Supplemental Data. Oshima et al. (2013). Plant Cell 10.1105/tpc.113.110783
18 / 18
MYB16r GAACATCGGTGAATCCGACGGTGAAG
GATC
quantitative RT-PCR for MYB16
PP2AA3f GACCAAGTGAACCAGGTTATTGG RT-PCR for PP2AA3
PP2AA3r TACTCTCCAGTGCCTGTCTTCA RT-PCR for PP2AA3
Supplemental reference 1. Reference for Supplemental Figure 2B
Obayashi, T., Hayashi, S., Saeki, M., Ohta, H., and Kinoshita, K. (2009). ATTED-II
provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37, 987-991.