Segmenting dna sequence into words
-
Upload
liang-wang -
Category
Technology
-
view
631 -
download
0
Transcript of Segmenting dna sequence into words

OUTLINE
1. Why we need ‘word’ sequence2 How to build DNA vocabulary2. How to build DNA vocabulary3. DNA sequence segmentationq g4. Some applications

1 WHY WE NEED ‘WORD’ SEQUENCE
Letter sequence: “hellowordiloveyou”, It’s means nothing. gwe need “hello world I love you”, a word sequenceword sequence.So do for computer!

WE NEED WORDS!English words are naturally segmented by space. For some languages like Chinese. No space. No
li it d b d iexplicit word boundaries.We need the “words” for building efficient information retrieval system natural language information retrieval system, natural language understanding, etc.

SEGMENTATION RESEARCH
What’s the segment?Convert the letter sequence into “words” sequence.“helloworld” to “hello world”. We add space or other delimiter to ‘segment’ the letter sequenceother delimiter to segment the letter sequence.Segmentation is key step for most Chinese Information Processing (CIP) systemsInformation Processing (CIP) systems.

So for “ATCCATTCCAGGCCAGGG……”?
If we could segment DNA sequence, we could:1. Apply many mature research like web search
engine into DNA analyzing.2. Get new tips for DNA function research.

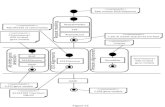
Two step for segment:1 Build word list or vocabulary.1. Build word list or vocabulary.2. Segment sequence based on this
b lvocabulary.3. Step 1 is key.p y

2 HOW TO BUILD DNA VOCABULARY?
Although we have many many DNA sequencesqWe still almost have no idea for it.T f li i ti k l dToo few linguistic knowledge……….So what?

Rosette stone

Rosette stone of DNA,still not found………..We only have many “Hieroglyphic text”.
Cracked it? The answer is YES!

Unsupervised segment research:

Unsupervised method: evaluate all possibleword’s probability.If k th d d th iIf we know the words and theirprobabilities …,we can get the segmented text.

Some unsupervised method to build vocabulary:
1. Frequency based method.2. Using n-gram language model.3. EM methods.

Frequency method:Probability of word: P(word) = C(word)/C(N)C(word) is number of word appear in corpus, C(N) is all word numbers.f l “ h i h ” for example: “who is who”. C(N)=3,C(who)=2,C(is)=1.S P( h ) 2/3 P(i ) 1/3So P(who)=2/3, P(is)=1/3
For 2-gram words.C( h i ) 1 C(i h ) 1 C(N) 2C(who is)=1,C(is who)=1,C(N)=2.So P(who is)=1/2,P(is who)=1/2

N-gram language model method:For 1-gram word, it’s same to frequency method.For n-grams word, n>2, for example:
P(who am i)=P(who)P(am|who)P(i|who am)Here,P(B|A)=C(AB)/C(A)

EM th dEM methods:1. For each sentence in the unsegmented text,
C t th lik lih d f h ibl Compute the likelihood of each possible segmentation using the current estimated values of the word probabilities.pThe segmentation likelihood is normalized as fraction“ that sums to 1.Count the words in each segmentation. I.e., add the fraction" of the segmentation to the word countcount.
2. Update the word probabilities using the word counts.
3. Repeat until convergence.

Apply to DNA:Select experiment data(full genomes):
AspergillusSchizosaccharomycesAcyrthosiphonZebrafish………………..

Before using unsupervised method. We need a important parameter: maximal word length.
U i f’ l t l t1. Use zipf’s laws to evaluate.2. Use language model to evaluate.

zipf’s laws: in a long enough document, about 50% words only occur once such word named 50% words only occur once, such word named “Hapax legomenon”.Assume the DNA word length is 1,2,……, then g , , ,calculate the percentage of “Hapax legomenon” respectively.O l i t t b ild h d F Overlapping segment to build such words. For example, “ATCAG”, for 3 word length, we get words “ATA”, “TCA”,”CAG”., ,If for a length, its percentage of “Hapaxlegomenon” is 50%, we use this length as word l th length.

0 8
0.9
1
0.5
0.6
0.7
0.8
0.2
0.3
0.4
9 10 11 12 13 14 15 160
0.1
For of most genomes, 50% line of ‘Hapaxlegomenon’ corresponding to word length 12 to 15

N-gram language model method:Assume DNA word length is 1,2,……, then calculate the language perplexities of sequence.Language perplexities describe the probability of all sequence.p o a y o a seq e ce.The lowest point of language perplexity will correspond to the maximal words will correspond to the maximal words length.

5
5.5
4.5
3.5
4
3
ld fi d th l l iti d ith
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 152.5
we could find the language perplexities reduce with the increase of word length n, till n<14. So for DNA sequence, Max word length is about 12-15q , g

We use 12bps as the maximal DNA word length.If Word length is n, 4^n parameters to be
l t d evaluated. Longer word length, represent more things . But We need much more DNA sequencesWe need much more DNA sequences.Word is a relative concept. We only need select a appropriate word lengthappropriate word length.See the research in collocation mining. “Ilovethebigapple” to “I love thebigapple” is Ilovethebigapple to I love thebigapple is better, “I love the big apple” is also ok.

Having the maximal word length.We could easy evaluate the probabilities of all possible DNA word by unsupervised methods mentioned above.For 12 word length, we get 4^1+4^2+….+4^12 = 22,369,620 words.…. ,369,6 0 wo s.All these words should be added into vocabulary? NOvocabulary? NO.

Filter the word list :1. Word frequency. Low occurrence word should
b d l t dbe deleted.2. MI feature. The connection of letters in word
should be strong enough should be strong enough. 3. Boundary Entropy feature. The “word” should
have clear boundary have clear boundary. 4. Other features, selectional association,
symmetric conditional probability Dice formula symmetric conditional probability, Dice formula, etc.

We mix all experimental data to train a DNA vocabulary. Aft filt i h l d t t b t After filtering whole word set, we get about 564,145 words. We use this words set as our “DNA vocabulary”DNA vocabulary .Having a “DNA vocabulary” with words probabilities. probabilities. Segment DNA sequence into “DNA words” is a easy mission.y

3 DNA SEQUENCE SEGMENTATION
Having a vocabulary with words probability. How to segment the sequence?F l ‘AGC’ ld b di id d i t ‘A /G For example: ‘AGC’ could be divided into, ‘A /G /C’, ‘AG /C’, ‘A /GC’,’AGC’.
Maximal probability segmentation method.S l t t ti f h i th i l 1. Select a segmentation form having the maximal probability as its segmentation.
2 Applying Dynamical programming method to 2. Applying Dynamical programming method to get this segmentation .

Metrics for segmentation. Precision? We have no preliminary knowledge for DNA dDNA words.Stability metrics for DNA segment:
S b d l l f h d 1. Sub sequence: delete some letters from head or tail of the original sequence.A d t ti th d h ld th 2. A good segmentation method should ensure the sub sequence is segmented into the same form with the original sequencewith the original sequence.
3. Stability :Calculate the percentage of same segmenting words between sub sequence and g g qoriginal sequence.

Vocabulary built by mixed experimental genome data. Segment different sequence:
genomes Acyrthosiphon Arabidopsis Aspergillus Caenorhabditis Zebrafish Fruit Fly
stability 0.942446 0.953038 0.949611 0.933767 0.904238 0.93521
genomes Human Mouse Oryza Schizosaccharomyces Strongylocentrotus Xenopus
stability 0.914045 0.898843 0.909858 0.957075 0.919044 0.92456
Build vocabulary by different genomes ,and segment corresponding sequence:
genomes Acyrthosiphon Arabidopsis Aspergillus Caenorhabditis Zebrafish Fruit Fly
stability 0.980074 0.986467 0.973245 0.98359 0.963535 0.983323
genomes Human Mouse Oryza Schizosaccharomyces Strongylocentrotus Xenopus
stability 0.974546 0.965113 0.969982 0.983754 0.970433 0.973462

For table above:Build a vocabulary by merged data of different
S t diff t t bilit genomes. Segment different sequences. stability > 93%.Building vocabulary by human genomes: Building vocabulary by human genomes: Segment sequence in human. Stability: > 95%. Segment sequence in rice or other genomes, Segment sequence in rice or other genomes, stability > 90%.

An interesting question : All genomes use the same language?di t1 b ilt b i di t 2 b ilt b h dict1:built by rice genome; dict 2 , built by human genome.Segment same sequence If two dicts segment it Segment same sequence. If two dicts segment it into same segmented form, they may use the same language!same language!Like segment stability metric.
1 Use two dictionary to segment one sequence 1. Use two dictionary to segment one sequence. Get two segmented sequences.
2. Calculate the percentage of same segmenting p g g gwords between two segmented sequences.

Build vocabulary by different chromosomes of human, segment same sequence. Its ‘stability ’ : about 85%about 85%.Build vocabulary by different genomes, segment same sequence This ‘stability ’ : about 35%--50%same sequence. This stability : about 35% 50%.

Why?Data sparse problem: some words only appear
l ti it b bilit i t li bl several times, its probability is not reliable. solution:
1 More sequences/corpus Single genome data is 1. More sequences/corpus. Single genome data is not enough to evaluate all word prob.
2 More smooth methods Reduce the word length 2. More smooth methods . Reduce the word length or filter more words will increase such stability.
3 This result shows: Different genomes is 3. This result shows: Different genomes is likely to use same language.

4 SOME APPLICATIONS
After segmenting ,almost all current text information processing p gtechnology could be directly applied in DNA analyzingin DNA analyzing.Using the dictionary built by mixed
d genomes data.

Hot topic( LDA method):The hot topics in different genomes:

Alignment:1. Current: compare letter by letter. 2. After segmenting, word by words, faster 3 We build a DNA search engine like 3. We build a DNA search engine like
Google.www dnasearchengine comwww.dnasearchengine.com

More application:DNA sequencing error : Automatic
f di proofreading. Genomes comparing: Plagiarize detecting.………

Thanks!Open source :https://code.google.com/p/dnasearchengine/