Protein sequencing presentation
-
Upload
mobin-aslam -
Category
Education
-
view
1.445 -
download
3
Transcript of Protein sequencing presentation

PROTEIN SEQUENCING
BYMOBIN ASLAM

• Biomolecules• Polymers of amino acids• Variation in protein structure and function is
due to the difference in amino acid sequence in peptide chains
PROTEIN

Protein sequencing
• Technique to find out the sequence of amino acids in a protein
Sequencing methods1-N-terminal sequencing (Edman degradation)2-C-terminal sequencing3-Prediction from DNA sequence

Edman degradation
N-terminal sequencing

STEPS
• Protein purification• Protein denaturation• Protein digestion• N-terminal labeling• Separation of labeled amino acid by
chromatography• Detection through mass spectrometry• Data analysis

Protein isolation(purification)
• 1-SDS-PAGE(sodium dodecyl sulfate-poly
acryl amide gel)2-Two dimensional gels
Protein of interest is immobilized by being absorbed onto a chemically modified glass or by electro blotting onto a porous polyvinylidene fluoride (PVDF) membrane.

by heating a sample of the protein in 6 Molar HCL up to 100-110 degrees Celsius for 24 hours or longer
It may degrade some amino acids
To avoid thisThiol reagents or phenol are
used- Performic acid for intra
chain or inter chain S-S bonds
Protein hydrolysis(denaturation)

Protein digestion
• Use Endoproteinase Lys-C, CNBr, Pepsin or trypsin to digest proteins into a population of peptides
• Other enzymes include Glu-C and chymotrypsin
• Add enzyme at 1:20 enzyme: protein ratio • incubate at room temperature for 6-9hrs• For better results use mixture of enzymes


N-terminal labeling
• The Edman reagent, phenylisothiocyanate (PTC), is added to the adsorbed peptide, together with a mildly basic buffer solution of 12% trimethylamine
• This reacts with the amine group of the N-terminal amino acid
• The terminal amino acid can then be selectively detached by the addition of anhydrous acid
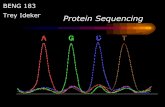
• The derivative then isomerises to give a substituted phenylthiohydantoin which can be washed off and identified by chromatography, and the cycle can be repeated


CHROMATOGRAPHY• Chromatography is a
technique in which molecules are separated based on volatility and bond characteristics when subjected to a carrier
• Derivatives of amino acid can be separated by
• 1-HPLC• 2-Gas chromatography• In gas chromatography (GC),
the mobile phase is an inert gas such as helium

MASS SPECTROMETERY
• Mass spectrometry (MS) is an analytical technique that measures the mass-to-charge ratio of charged particles
• The MS principle consists of ionizing chemical compounds to generate charged molecules or molecule fragments and measuring their mass-to-charge ratios
• Separated amino acid derivatives are analyzed by mass spectrometer

MS procedure
• A sample is loaded onto the MS instrument, and undergoes vaporization
• The components of the sample are ionized by one of a variety of methods (e.g., by impacting them with an electron beam), which results in the formation of charged particles (ions)
• The ions are separated according to their mass-to-charge ratio in an analyzer by electromagnetic fields
• The ions are detected, usually by a quantitative method • The ion signal is processed into mass spectra

Mass spectrometer

• first strategy for identifying an unknown compound is to compare its experimental mass spectrum against a library of mass spectra
• Standard solutions of amino acids are also used and the resulting pattern is compared with standard spectrum.
MS data analysis

Limitations of Edman degradation
• Need Pure Samples of Peptides• Requires 40-60 min / Amino Acid• Can’t Analyze N-Terminally Modified Peptides
• Advantages• Most Reliable Sequencing Technique

C-terminal sequencing
• The number of methods available for C-terminal amino acid analysis is much smaller than the number of available methods of N-terminal analysis. The most common method is to add carboxypeptidases to a solution of the protein, take samples at regular intervals, and determine the terminal amino acid by analyzing a plot of amino acid concentrations against time.

Prediction from DNA sequence
• Protein sequence can also be determined indirectly from the mRNA or, in organisms that do not have introns (e.g. prokaryotes)
• Sequence a short section, perhaps 15 amino acids long, of the protein
• Design primers from the amino acid sequence and amplify the gene, sequence the gene and determine the amino acid sequence of protein

Automatic protein sequencers
• Automatic protein sequencers are designed that perform the 3 steps(labeling, separation and analysis of amino acids) at a time and analyze data and give results automatically

Applications of protein sequencing
• Recombinant protein synthesis• Drugs production• Antibiotic production• Functional genomics• Determine the protein folding patterns• In bioinformatics• It plays vital role in proteomics• Used for the prediction of final structure, function and
location of protein• To find out the location of gene coding for that protein

???
THANKS