Nat. Struct. Mol. Biol. (10 July 2011) doi:10.1038/nsmb ... · Mol. Biol. (10 July 2011)...
Transcript of Nat. Struct. Mol. Biol. (10 July 2011) doi:10.1038/nsmb ... · Mol. Biol. (10 July 2011)...

nature structural & molecular biology
Nat. Struct. Mol. Biol. (10 July 2011) doi:10.1038/nsmb.2100
SUMOylation regulates telomere length homeostasis by targeting Cdc13Lisa E Hang, Xianpeng Liu, Iris Cheung, Yan Yang & Xiaolan Zhao
In the version of this supplementary file originally posted online on 10 July 2011, a formatting error caused numerous lines to disappear. The errors have been corrected in this file as of 28 July 2011.
C O R R E C T I O N N OT I C E

Sumoylation regulates telomere length homeostasis by targeting Cdc13
Lisa E. Hang, Xianpeng Liu, Iris Cheung, Yan Yang, and Xiaolan Zhao
Supplementary Materials. Supplementary Figures 1-6. Supplementary Table 1 References

a Rap1‐MycWT siz∆
Yku80WT siz∆
WTsiz1∆ siz2∆ siz1∆ siz2∆
mms21 siz2∆ WT
e
ProAorMyc
SUMO
ProA
SUMO
Cdc13
Pif1WT mms21
b
ProA
SUMO
Supplementary Figure 1 Sumoylation of telomere proteins is differentially regulated by three SUMO E3s and sumoylation of Cdc13 is primarily dependent on Siz1 and Siz2. (a,b,d) Siz and Mms21 SUMO E3s affect the sumoylation of telomere proteins differently. Sumoylation of Cdc13, Yku80 and Rap1 is greatly reduced in siz1∆ siz2∆ (siz∆) cells (a), but not in mms21-11 (mms21) cells (d), while sumoylation of Pif1 is reduced in mms21-11 cells as evidenced by a decrease in the ratio of sumoylated to unmodified Pif1 (b). (c) Cdc13 sumoylation is demonstrated using His6-Flag-SUMO. The His6-Flag (HF) module added to SUMO (Smt3) causes a shift of sumoylated Cdc13. Cells contain either Cdc13-TAP (Top) and Cdc13-Myc (Bottom). Untagged Cdc13 and cdc13-snm-TAP are included as controls. (e) Sumoylation of Cdc13 is greatly diminished in siz1∆ siz2∆, but not in siz1∆, siz2∆, or mms21-CH siz2∆ (mms21 siz2∆) mutants. All experiments were performed as in Figure 1d. Proteins tagged with Myc are indicated and all others are tagged with TAP.
dCdc13
WT mms21
Rap1‐MycWT mms21
Yku80‐MycWT mms21
SUMO
ProAorMyc
untagged
c
Cdc13‐TAP
WT WT snm
HF‐Smt3Smt3
Smt3
Cdc13: WT
Cdc13‐mycuntagged
ProA
SUMO
Myc
SUMO
HF‐Smt3
Cdc13WT siz∆

50
0 15 30 45 75 90 105 60 (Min)
Enr
ichm
ent
0
10
20
30
40
Est1-Myc Tel XVL
0 15 30 45 75 90 105 60 (Min)
Enr
ichm
ent
0
10
20
30
40
Est1-Myc Tel VIR
50
S phase S phase
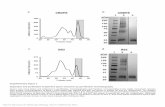
Supplementary Figure 2 Est1 is enriched at telomeres in late S phase. ChIP analysis of Est1 binding at two telomeres, Tel XVL (left) and Tel VIR (right), is shown at different time points after G1 cells (0 min) were released into the cell cycle.

Wild‐type
G115’30’45’60’75’
90’105’
a b
cdc13‐snm
G115’30’45’60’75’90’105’
1.5
2.5
3
4
56
10
kb
c d
1
1.5
2
2.5
3
4
1
2
3
4
5
WT
cdc13‐snm
cdc13‐snm est1∆
est1∆
kb
Supplementary Figure 3 cdc13-snm increases telomere length in YPH strain background, but does not affect cell cycle progression or senescence. (a) Wild-type and cdc13-snm cells were arrested in G1 phase by alpha-factor and released into the cell cycle. FACS analysis was performed at the indicated time points. (b) cdc13-snm in YPH background exhibits longer telomeres than isogenic wild-type cells. (c,d) same as Figure 3b,c, except est1∆ was used, instead of tlc1∆.

Denatured
WT snm CDC13: snm WT
30°C 37°Ca
NaTve
1
1.5
2
0.5
3
4
56
kb
Supplementary Figure 4 cdc13-snm does not affect G-overhangs, and siz1∆ siz2∆ is epistatic with cdc13-snm and weakens the Cdc13–Stn1 interaction. (a) The levels of G-overhangs in cdc13-snm cells are not altered at 30° C and 37° C. DNA fragments containing G-overhangs are indicated by arrows. (b) The telomere length of siz1∆ siz2∆ cdc13-snm cells does not extend beyond those of either cdc13-snm or siz1∆ siz2∆ mutants. (c) The Cdc13–Stn1, but not the Rtt107–Slx4, interaction is reduced in siz1∆ siz2∆ two-hybrid cells. Wild-type and siz1∆ siz2∆(siz∆) two-hybrid strains containing pairs of indicated GBD and GAD plasmids were grown on medium lacking leucine and tryptophan (-LEU-TRP), and the activation of the ADE reporter was scored by replica plating onto -LEU-TRP-ADE media as in Figure 4a.
BasepairincreaseoverWT:
1
0.75
1.5
2
2.5
+55 +60 +60 0
Kb
siz∆
pOADpOBD
RY107 Slx4
vec
Cdc13
Cdc13
vec
Stn1
Stn1
‐LEU‐TRP ‐LEU‐TRP‐ADE
WTstrains
plasmids siz∆ WT
b c

30°C 37°C
a
b
yku80∆ cdc13‐snm
cdc13‐snm
yku80∆
WT
yku80∆
Supplementary Figure 5 cdc13-snm rescues the temperature sensitivity of yku70∆ and yku80∆ without affecting the levels of G-overhangs. (a,b) cdc13-snm rescues the lethality of yku70∆ (a) and yku80∆ (b) cells at 37° C. (c) G-overhang levels in yku70∆ cells are not affected by cdc13-snm at 30° C. DNA fragments containing G-overhangs are indicated by arrows.
yku70∆ cdc13‐snm yku70∆
Denatured
NaTve
c
1
1.5
2
0.5
3
4
5
6
kb
WT
cdc13‐snm
yku70∆
cdc13‐snm yku70∆
30°C 37°C

+
BasepairincreaseoverWT:
Stn1O/E:WT snm CDC13: – –+
1
1.5
2.0
2.5
0.75
0 ‐55+55‐20
kb
Supplementary Figure 6 Stn1 overexpression suppresses the telomere length defect of cdc13-snm. Wild-type and cdc13-snm cells were transformed with either an Stn1 overexpression plasmid (+) or a control vector (–).

Supplementary Table 1. Strains and Plasmids Name Genotype Sources W1588-4C MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 R. Rothstein JBY491 MATα ubc9∆::TRP1 leu2-3,112-ubc9-1::LEU2 ref. 5 T346-1B UBC9-13MYC::HIS3 this study T357-1B UBC9-3HA::KAN this study Z364-4B siz1∆::KAN siz2∆::URA3 cir0 this study Z357-1B siz1∆::KAN mms21-CH::HIS3 cir0 this study Z357-1A siz2∆::URA3 mms21-CH::HIS3 cir0 this study Z375-4B siz1∆::KAN cir0 this study Z357-1C siz2∆::URA3 cir0 this study Z357-2D mms21-CH::HIS3 cir0 this study T240-9D RAP1-13MYC::HIS5 this study T439-2 MATa YKU70-ProA-KAN this study T496-4 MATα CDC13-TAP::HIS3 this study G760-1 MATa PIF1-13MYC::TRP bar1Δ::HIS3 Zakian lab T238-22D MATα YKU80-13MYC-HIS5 ref. 1 X3299-1D YKU70-13MYC::HIS3 this study G532 MATa PIF1-TAP::HIS3 in S288C ref. 2 X591-7A RAD52-TAP::HIS5 this study T495-1 MATα cdc13-K909R-TAP::HIS3 this study X3593-1B yku70∆::LEU2 cdc13-K909R-TAP::HIS3 this study X1475-7D yku70∆::LEU2 this study X3302-2A yku70∆::LEU2 CDC13-TAP::HIS3 this study IC1-8H CDC13/cdc13-snm TLC1/tlc1∆::HIS3 this study
pJ69-4a MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4∆ gal80∆ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ ref. 3
pXZ389 pOBD-CDC13 this study pXZ390 pOBD-cdc13-snm this study pXZ412 pOAD-STN1 this study p168 pGADT7-POL1N ref. 4 X3216-3C tel1∆::URA3 this study X3216-3D tel1∆::URA3 cdc13-K909R-TAP::HIS3 this study X3258-1C CDC13-TAP::HIS3 TRP1::pGAL-13MYC-EST1 this study X3259-3D cdc13-K909R-TAP::HIS3 TRP1::pGAL-13MYC-EST1 this study X3215-1C CDC13-TAP::HIS3 tel1∆::URA3 this study X3260-6D TRP1::pGal-13MYC-STN1 CDC13-TAP::HIS3 this study X3261-2B TRP1::pGal-13MYC-STN1 cdc13-K909R-TAP::HIS3 this study X3263-5B stn1∆C199::KAN this study X3263-1D stn1∆C199::KAN cdc13-K909R-TAP::HIS3 this study X3262-10A CDC13-TAP::HIS3 stn1∆C199::KAN this study

X3301-2D RAP1-13MYC::HIS3 stn1∆C199::KAN this study X3299-1A YKU70-13MYC::HIS5 stn1∆C199::KAN this study X3188-2A CDC13-TAP::HIS3 rif1∆::URA3 this study T856-2 CDC13-SMT3-3HA::KAN this study T1036-1 CDC13-T308A-3HA::KAN this study T1021-2 CDC13-T308A-SMT3-3HA::KAN this study T881-2 TRP1::pGal-13MYC-STN1 CDC13-3HA::KAN this study X3326-10A TRP1::pGal-13MYC-STN1 CDC13-SMT3-3HA::KAN this study T880-2 CDC13-3HA::KAN this study
EHB10355 MATa bar1Δ::NAT or bar1Δ::LEU2 ura3 trp1 leu2 CDC13(T308A)-3HA::KAN in A364a ref. 6
T882-1 CDC13-K909R-3HA::KAN this study T855-4 CDC13-T308A, K909R-TAP::HIS3 this study T854 CDC13-T308A-TAP::HIS3 this study X1952-31A YKU80-13MYC::HIS5 siz1∆::KAN siz2∆::URA3 this study X1951-1C RAP1-13MYC::HIS5 siz1∆::KAN siz2∆::URA3 this study Z365-2A CDC13-TAP::HIS3 siz1∆::KAN siz2∆::URA3 cir0 this study T238-24D YKU80-13MYC::HIS5 mms21-11::LEU2 this study T240-2D RAP1-13MYC::HIS3 mms21-11::URA3 this study X1515-5D CDC13-TAP::HIS3 mms21-11::URA3 this study X1599-4B PIF1-TAP::HIS3 mms21-11::URA3 this study Z365-2D CDC13-TAP::HIS3 siz1Δ::KAN cir0 this study Z365-4B CDC13-TAP::HIS3 siz2Δ::URA3 cir0 this study X3710-12B CDC13-TAP::HIS3 mms21-CH::HIS siz2Δ::URA3 this study X3579-2B CDC13-TAP::HIS3 HF-SMT3::LEU2 this study X3612-7C CDC13-K909R-TAP::HIS3 HF-SMT3::LEU2 this study T1041-3 CDC13-3MYC::TRP1 this study X3709-1C CDC13-3MYC::TRP1 HF-SMT3::LEU2 this study T699-1 CDC13::HIS5 in YPH499 this study T713 cdc13-snm::HIS5 in YPH499 this study IC1-8I CDC13/cdc13-snm EST1/est1∆::HIS3 this study X1770-19C yku80∆::URA3 this study X1563-1B yku80∆::URA3 cdc13-K909R-TAP::HIS3 this study X1770-19D yku80∆::URA3 CDC13-TAP::HIS3 this study Z364-2C cdc13-snm-TAP::HIS siz1Δ::KAN siz2Δ::URA3 cir0 this study
G701 MATa ura3052, lys2-801 amber, ade2-101 ochre, trp1-∆63 his3-∆200, leu2-∆1 in YPH499 Zakian lab
G625 cdc13-snm in YPH499 Zakian lab Strains in this study are derivatives of W1588-4C, a RAD5 derivative of W303 (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1 rad5-535 7, unless noted otherwise. A single representative of each genotype is listed. Stn1 overexpression plasmids and control vectors are described in ref. 6.

References
1 Zhao, X. & Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102, 4777-4782 (2005).
2 Huh, W. K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686-691 (2003).
3 James, P., Halladay, J. & Craig, E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436 (1996).
4 Qi, H. & Zakian, V. A. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev. 14, 1777-1788 (2000).
5 Bachant, J., Alcasabas, A., Blat, Y., Kleckner, N. & Elledge, S. J. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9, 1169-1182 (2002).
6 Li, S. et al. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136, 50-61 (2009).
7 Thomas, B. J. & Rothstein, R. Elevated recombination rates in transcriptionally active DNA. Cell 56, 619-630 (1989).