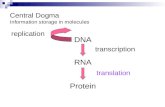
RNAi Mechanism. The Central Dogma DNA (double-stranded) RNA (single-stranded) Protein.
Flow of Genetic Information The central dogma is the concept that cells are governed by a cellular...
-
Upload
shawn-evans -
Category
Documents
-
view
217 -
download
0
Transcript of Flow of Genetic Information The central dogma is the concept that cells are governed by a cellular...
Flow of Genetic Information
• The central dogma is the concept that cells are governed by a cellular chain of command:
DNA RNA protein
© 2011 Pearson Education, Inc.
Figure 17.3
DNA
mRNARibosome
Polypeptide
TRANSCRIPTION
TRANSLATION
TRANSCRIPTION
TRANSLATION
Polypeptide
Ribosome
DNA
mRNA
Pre-mRNARNA PROCESSING
(a) Bacterial cell (b) Eukaryotic cell
Nuclearenvelope
Figure 17.4
DNAtemplatestrand
TRANSCRIPTION
mRNA
TRANSLATION
Protein
Amino acid
Codon
Trp Phe Gly
5
5
Ser
U U U U U3
3
53
G
G
G G C C
T
C
A
A
AAAAA
T T T T
T
G
G G G
C C C G G
DNAmolecule
Gene 1
Gene 2
Gene 3
C C
Cracking the Genetic Code• 20 amino acids but there are only four nucleotide
bases in DNA• All 64 codons ( 43) were deciphered by the mid-
1960s• Of the 64 triplets, 61 code for amino acids; 3
triplets are “stop” signals to end translation• The genetic code is redundant (more than one
codon may specify a particular amino acid) but not ambiguous; no codon specifies more than one amino acid
© 2011 Pearson Education, Inc.
Figure 17.5Second mRNA base
Fir
st m
RN
A b
ase
(5 e
nd
of
cod
on
)
Th
ird
mR
NA
bas
e (3
en
d o
f co
do
n)
UUU
UUC
UUA
CUU
CUC
CUA
CUG
Phe
Leu
Leu
Ile
UCU
UCC
UCA
UCG
Ser
CCU
CCC
CCA
CCG
UAU
UACTyr
Pro
Thr
UAA Stop
UAG Stop
UGA Stop
UGU
UGCCys
UGG Trp
GC
U
U
C
A
U
U
C
C
CA
U
A
A
A
G
G
His
Gln
Asn
Lys
Asp
CAU CGU
CAC
CAA
CAG
CGC
CGA
CGG
G
AUU
AUC
AUA
ACU
ACC
ACA
AAU
AAC
AAA
AGU
AGC
AGA
Arg
Ser
Arg
Gly
ACGAUG AAG AGG
GUU
GUC
GUA
GUG
GCU
GCC
GCA
GCG
GAU
GAC
GAA
GAG
Val Ala
GGU
GGC
GGA
GGGGlu
Gly
G
U
C
A
Met orstart
UUG
G
Transcription (DNA to mRNA)• RNA synthesis is catalyzed by RNA polymerase,
which prys the DNA strands apart and hooks together the RNA nucleotides
• DNA sequence where RNA polymerase attaches is called the promoter; in bacteria, the sequence signaling the end of transcription is called the terminator
© 2011 Pearson Education, Inc.
Stage 1: Initiation• Promoters signal the transcriptional start point
and usually extend several dozen nucleotide pairs upstream of the start point
• Transcription factors mediate the binding of RNA polymerase and the initiation of transcription
• The completed assembly of transcription factors and RNA polymerase II bound to a promoter is called a transcription initiation complex
• A promoter called a TATA box is crucial in forming the initiation complex in eukaryotes
© 2011 Pearson Education, Inc.
Figure 17.8
Transcription initiationcomplex forms
3
DNAPromoter
Nontemplate strand
53
53
53
Transcriptionfactors
RNA polymerase II
Transcription factors
53
53
53
RNA transcript
Transcription initiation complex
5 3
TATA box
T
T T T T T
A A A AA
A A
T
Several transcriptionfactors bind to DNA
2
A eukaryotic promoter1
Start point Template strand
Stage 2: Elongation
• As RNA polymerase moves along the DNA, it untwists the double helix, 10 to 20 bases at a time
• Transcription progresses at a rate of 40 nucleotides per second in eukaryotes in the 5’ to 3’ direction on growing RNA molecule
• A gene can be transcribed simultaneously by several RNA polymerases
© 2011 Pearson Education, Inc.
Nontemplatestrand of DNA
RNA nucleotides
RNApolymerase
Templatestrand of DNA
3
35
5
5
3
Newly madeRNA
Direction of transcription
A
A A A
AA
A
T
TT
T
TTT G
GG
C
C C
CC
G
C CC A AA
U
U
U
end
Figure 17.9
Stage 3: Termination
• The mechanisms of termination are different in bacteria and eukaryotes
• In bacteria, the polymerase stops transcription at the end of the terminator and the mRNA can be translated without further modification
• In eukaryotes, RNA polymerase II transcribes the polyadenylation signal sequence
© 2011 Pearson Education, Inc.
Eukaryotic cells modify RNA after transcription with enzymes
© 2011 Pearson Education, Inc.
• Modify ends• Each end of a pre-mRNA molecule is
modified with the 5 end receiving a modified nucleotide 5 cap and the 3 end gets a poly-A tail
• Modifications facilitate the export of mRNA to the cytoplasm, protect mRNA from hydrolytic enzymes and help ribosomes attach to the 5 end
• Splice out noncoding regions
Figure 17.10
Protein-codingsegment
Polyadenylationsignal
5 3
35 5Cap UTRStartcodon
G P P P
Stopcodon
UTR
AAUAAA
Poly-A tail
AAA AAA…
Split Genes and RNA Splicing
• Most eukaryotic genes and their RNA transcripts have long noncoding stretches of nucleotides that lie between coding regions called introns
• The other regions are called exons because they are eventually expressed, usually translated into amino acid sequences
• RNA splicing uses splicesomes to recognize small nuclear ribonucleoproteins (snRNPs) and removes introns and joins exons, creating an mRNA molecule with a continuous coding sequence
© 2011 Pearson Education, Inc.
Figure 17.11
5 Exon Intron Exon
5CapPre-mRNACodonnumbers
130 31104
mRNA 5Cap
5
Intron Exon
3 UTR
Introns cut out andexons spliced together
3
105 146
Poly-A tail
Codingsegment
Poly-A tail
UTR1146
Figure 17.12-3RNA transcript (pre-mRNA)
5Exon 1
Protein
snRNA
snRNPs
Intron Exon 2
Other proteins
Spliceosome
5
Spliceosomecomponents
Cut-outintronmRNA
5Exon 1 Exon 2
Evolutionary/Functional Importance of Introns
• Some introns contain sequences that may regulate gene expression
• Some genes can encode more than one kind of polypeptide, depending on which segments are treated as exons during splicing
• This is called alternative RNA splicing• Consequently, the number of different proteins an
organism can produce is much greater than its number of genes
• Exon shuffling may result in the evolution of new proteins
© 2011 Pearson Education, Inc.
GeneDNA
Exon 1 Exon 2 Exon 3Intron Intron
Transcription
RNA processing
Translation
Domain 3
Domain 2
Domain 1
Polypeptide
Figure 17.13
Translation ( mRNA to proteins
• tRNAs transfer amino acids to the growing polypeptide in a ribosome
• Molecules of tRNA are not identical– Each carries a specific amino acid on one end
– Each has an anticodon on the other end; the anticodon base-pairs with a complementary codon on mRNA
• Flattened into one plane to reveal its base pairing, a tRNA molecule looks like a cloverleaf
© 2011 Pearson Education, Inc.
Figure 17.14
Polypeptide
Ribosome
Trp
Phe Gly
tRNA withamino acidattached
Aminoacids
tRNA
Anticodon
Codons
U U U UG G G G C
AC C
C
CG
A A A
CGC
G
5 3mRNA
Figure 17.15
Amino acidattachmentsite
3
5
Hydrogenbonds
Anticodon
(a) Two-dimensional structure (b) Three-dimensional structure(c) Symbol used
in this book
Anticodon Anticodon3 5
Hydrogenbonds
Amino acidattachmentsite5
3
A A G
• Accurate translation requires two steps– First: a correct match between a tRNA and an
amino acid, done by the enzyme aminoacyl-tRNA synthetase
– Second: a correct match between the tRNA anticodon and an mRNA codon
• Flexible pairing at the third base of a codon is called wobble and allows some tRNAs to bind to more than one codon
© 2011 Pearson Education, Inc.
Aminoacyl-tRNAsynthetase (enzyme)
Amino acid
P P P Adenosine
ATP
P
P
P
PPi
i
i
Adenosine
tRNA
AdenosineP
tRNA
AMP
Computer model
Aminoacid
Aminoacyl-tRNAsynthetase
Aminoacyl tRNA(“charged tRNA”)
Figure 17.16-4
Ribosomes
• Ribosomes facilitate specific coupling of tRNA anticodons with mRNA codons in protein synthesis
• The two ribosomal subunits (large and small) are made of proteins and ribosomal RNA (rRNA)
• A ribosome has three binding sites for tRNA– The P site holds the tRNA that carries the growing
polypeptide chain
– The A site holds the tRNA that carries the next amino acid to be added to the chain
– The E site is the exit site, where discharged tRNAs leave the ribosome
© 2011 Pearson Education, Inc.
tRNAmolecules
Growingpolypeptide Exit tunnel
E PA
Largesubunit
Smallsubunit
mRNA5
3
(a) Computer model of functioning ribosome
Exit tunnel Amino end
A site (Aminoacyl-tRNA binding site)
Smallsubunit
Largesubunit
E P AmRNA
E
P site (Peptidyl-tRNAbinding site)
mRNAbinding site
(b) Schematic model showing binding sites
E site (Exit site)
(c) Schematic model with mRNA and tRNA
5 Codons
3
tRNA
Growing polypeptide
Next aminoacid to beadded topolypeptidechain
Figure 17.17
Building a Polypeptide-Stage 1:Initiation
• The initiation stage of translation brings together mRNA, a tRNA with the first amino acid, and the two ribosomal subunits
• First, a small ribosomal subunit binds with mRNA and a special initiator tRNA
• Then the small subunit moves along the mRNA until it reaches the start codon (AUG)
• Proteins called initiation factors bring in the large subunit that completes the translation initiation complex
© 2011 Pearson Education, Inc.
Figure 17.18
InitiatortRNA
mRNA
5
53Start codon
Smallribosomalsubunit
mRNA binding site
3
Translation initiation complex
5 33U
UA
A GC
P
P site
i
GTP GDP
Met Met
Largeribosomalsubunit
E A
5
Stage 2: Elongation of the Polypeptide
• During the elongation stage, amino acids are added one by one to the preceding amino acid
• Each addition involves proteins called elongation factors and occurs in three steps: codon recognition, peptide bond formation, and translocation
• Translation proceeds along the mRNA in a 5′ to 3′ direction
© 2011 Pearson Education, Inc.
Amino end ofpolypeptide
mRNA
5
E
Asite
3
E
GTP
GDP P i
P A
E
P A
GTP
GDP P i
P A
E
Ribosome ready fornext aminoacyl tRNA
Psite
Figure 17.19-4
Stage 3:Termination of Translation
• Termination occurs when a stop codon in the mRNA reaches the A site of the ribosome
• The A site accepts a protein called a release factor
• The release factor causes the addition of a water molecule instead of an amino acid
• This reaction releases the polypeptide, and the translation assembly then comes apart
© 2011 Pearson Education, Inc.
Figure 17.20-3
Releasefactor
Stop codon(UAG, UAA, or UGA)
3
5
3
5
Freepolypeptide
2 GTP
5
3
2 GDP 2 iP
Figure 17.21Completedpolypeptide
Incomingribosomalsubunits
Start ofmRNA(5 end)
End ofmRNA(3 end)(a)
Polyribosome
Ribosomes
mRNA
(b)0.1 m
Growingpolypeptides
Completing and Targeting the Functional Protein
• Polypeptide chains are modified after translation or targeted to specific sites in the cell
• During and after synthesis, a polypeptide chain spontaneously coils and folds into its three-dimensional shape
• Proteins may also require post-translational modifications before doing their job
• Some polypeptides are activated by enzymes that cleave them
• Other polypeptides come together to form the subunits of a protein
© 2011 Pearson Education, Inc.
Targeting Polypeptides to Specific Locations
• Two populations of ribosomes are evident in cells: free ribsomes (in the cytosol) and bound ribosomes (attached to the ER)
• Free ribosomes mostly synthesize proteins that function in the cytosol
• Bound ribosomes make proteins of the endomembrane system and proteins that are secreted from the cell
• Ribosomes are identical and can switch from free to bound
© 2011 Pearson Education, Inc.
Figure 17.22
Ribosome
mRNA
Signalpeptide
SRP
1
SRPreceptorprotein
Translocationcomplex
ERLUMEN
2
3
45
6
Signalpeptideremoved
CYTOSOL
Protein
ERmembrane
Mutations
• Mutations are changes in the genetic material of a cell or virus
• Point mutations are chemical changes in just one base pair of a gene
• The change of a single nucleotide in a DNA template strand can lead to the production of an abnormal protein
© 2011 Pearson Education, Inc.
Figure 17.23
Wild-type hemoglobin
Wild-type hemoglobin DNA3
3
35
5 3
35
5553
mRNA
A AGC T T
A AGmRNA
Normal hemoglobin
Glu
Sickle-cell hemoglobin
Val
AA
AUG
GT
T
Sickle-cell hemoglobin
Mutant hemoglobin DNAC
Types of Small-Scale Mutations
• Point mutations within a gene can be divided into two general categories
– Nucleotide-pair substitutions
– One or more nucleotide-pair insertions or deletions
© 2011 Pearson Education, Inc.
Substitutions
• A nucleotide-pair substitution replaces one nucleotide and its partner with another pair of nucleotides
• Silent mutations have no effect on the amino acid produced by a codon because of redundancy in the genetic code
• Missense mutations still code for an amino acid, but not the correct amino acid
• Nonsense mutations change an amino acid codon into a stop codon, nearly always leading to a nonfunctional protein
© 2011 Pearson Education, Inc.
Wild type
DNA template strand
mRNA5
5
3
Protein
Amino end
A instead of G
(a) Nucleotide-pair substitution
3
3
5
Met Lys Phe Gly StopCarboxyl end
T T T T T
TTTTTA A A A A
AAAACC
C
C
A
A A A A A
G G G G
GC C
G GGU U U U UG
(b) Nucleotide-pair insertion or deletion
Extra A
35
53
Extra U
5 3
T T T T
T T T T
A
A A A
A
AT G G G G
GAAA
AC
CCCC A
T35
5 3
5T T T T TAAAACCA AC C
TTTTTA A A A ATG G G G
U instead of C
Stop
UA A A A AG GGU U U U UG
MetLys Phe Gly
Silent (no effect on amino acid sequence)
T instead of C
T T T T TAAAACCA GT C
T A T T TAAAACCA GC C
A instead of G
CA A A A AG AGU U U U UG UA A A AG GGU U U G AC
AA U U A AU UGU G G C UA
GA U A U AA UGU G U U CG
Met Lys Phe Ser
Stop
Stop Met Lys
missing
missing
Frameshift causing immediate nonsense(1 nucleotide-pair insertion)
Frameshift causing extensive missense (1 nucleotide-pair deletion)
missing
T T T T TTCAACCA AC G
AGTTTA A A A ATG G G C
Leu Ala
Missense
A instead of T
TTTTTA A A A ACG G A G
A
CA U A A AG GGU U U U UG
TTTTTA T A A ACG G G G
Met
Nonsense
Stop
U instead of A
35
35
53
35
53
35 3Met Phe Gly
No frameshift, but one amino acid missing(3 nucleotide-pair deletion)
missing
35
53
5 3U
T CA AA CA TTAC G
TA G T T T G G A ATC
T T C
A A G
Met
3
T
A
Stop
35
53
5 3
Figure 17.24
Insertions and Deletions
• Insertions and deletions are additions or losses of nucleotide pairs in a gene
• These mutations have a disastrous effect on the resulting protein more often than substitutions do
• Insertion or deletion of nucleotides may alter the reading frame, producing a frameshift mutation
© 2011 Pearson Education, Inc.
Figure 17.24d
Wild type
DNA template strand
mRNA5
5
Protein
Amino endStopCarboxyl end
33
3
5
Met Lys Phe Gly
A
A
A A
A A A A
A AT
T T T T T
T T TT
C C C C
C
C
G G G G
G
G
A
A A A AG GGU U U U U
(b) Nucleotide-pair insertion or deletion: frameshift causingimmediate nonsense
Extra A
Extra U
53
5
3
3
5
Met
1 nucleotide-pair insertion
Stop
A C A A GT T A TC T A C G
T A T AT G T CT GG A T GA
A G U A U AU GAU G U U C
A T
A
AG
Figure 17.24e
DNA template strand
mRNA5
5
Protein
Amino endStopCarboxyl end
33
3
5
Met Lys Phe Gly
A
A
A A
A A A A
A AT
T T T T T
T T TT
C C C C
C
C
G G G G
G
G
A
A A A AG GGU U U U U
(b) Nucleotide-pair insertion or deletion: frameshift causingextensive missense
Wild type
missing
missing
A
U
A A AT T TC C A T TC C G
A AT T TG GA A ATCG G
A G A A GU U U C A AG G U 3
53
35
Met Lys Leu Ala
1 nucleotide-pair deletion
5
Figure 17.24f
DNA template strand
mRNA5
5
Protein
Amino endStopCarboxyl end
33
3
5
Met Lys Phe Gly
A
A
A A
A A A A
A AT
T T T T T
T T TT
C C C C
C
C
G G G G
G
G
A
A A A AG GGU U U U U
(b) Nucleotide-pair insertion or deletion: no frameshift, but oneamino acid missing
Wild type
AT C A A A A T TC C G
T T C missing
missing
Stop
53
35
35
Met Phe Gly
3 nucleotide-pair deletion
A GU C A AG GU U U U
T GA A AT T TT CG G
A A G
Mutagens
• Spontaneous mutations can occur during DNA replication, recombination, or repair
• Mutagens are physical or chemical agents that can cause mutations
© 2011 Pearson Education, Inc.
Comparing Gene Expression in Bacteria, Archaea, and Eukarya
• Bacteria and eukarya differ in their RNA polymerases, termination of transcription, and ribosomes; archaea tend to resemble eukarya in these respects
• Bacteria can simultaneously transcribe and translate the same gene
• In eukarya, transcription and translation are separated by the nuclear envelope
• In archaea, transcription and translation are likely coupled
© 2011 Pearson Education, Inc.
Figure 17.26TRANSCRIPTION
DNA
RNApolymerase
ExonRNAtranscript
RNAPROCESSING
NUCLEUS
Intron
RNA transcript(pre-mRNA)
Poly-A
Poly-A
Aminoacyl-tRNA synthetase
AMINO ACIDACTIVATION
Aminoacid
tRNA
5 C
ap
Poly-A
3
GrowingpolypeptidemRNA
Aminoacyl(charged)tRNA
Anticodon
Ribosomalsubunits
A
AE
TRANSLATION
5 Cap
CYTOPLASM
P
E
Codon
Ribosome
5
3