Fig. S 1
description
Transcript of Fig. S 1

Fig. S 1
a b
CJ-induced genes classed by ontology. Distribution of CJ induced genes, obtained by cDNA microarray (a) and ATH1 oligonucleotide microarray (b) analysis, into functional categories. Genes were assigned to various functional categories based on searching the TAIR or MIPS databases for suggested GO annotations.

control
At2g44130::LUX
+CJ
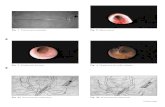
Fig. S 2
The promoter of the F-box At3g28740 was translationally fused to luciferase and stably introduced into Arabidopsis. Expression was induced by the exogenous cis-jasmone vapours (b) but not in the control (a). Luciferase activity was detected after application of luciferin and imaging of whole plants (rossette stage) via CCD. Expression was detected predominantly in the midrib of leaves.
a b

cis-jasmone inducible promoters at 20 hpt (cDNA array)
background (random Arabidopsis promoters)
cis-jasmone inducible promoters at 3 hpt (affymetrix array)
a b
ab b
YACGTGGC(ABRE core motif)
YACGTGGC(MYB core motif)
TGAC
TGACGT
YAACKG (myb2 motif)
RCCGAC (DRE/CRT core motif)
WRKY motif : TAGWCT(van der Ent et al., 2009)
Fig. S 3
background (random Arabidopsis promoters)
cis-jasmone inducible promoters at 3 and/or 20 hpt
Fig. S3: Occurrences of cis-acting elements in the promoter regions of CJ-inducible genes. motifs were quantified in the 500-bp sequences preceding the 5'-end of each transcription unit according to POBO bootstrapping analysis (Kankainen & Holm, 2004b). Different letters indicate statistically significant differences in occurrences of cis-elements between promoter groups (χ2 test; α=0.05).
a Occurrences of as-1 core motif TGAC in CJ-inducible genes at 3h and 20h of exposure to CJ in comparison to randomly selected promoter sequences from the Arabidopsis genome. b Occurrences of core sequences of different stress-related cis elements in the combined set of CJ-inducible genes at 3h and 20h of exposure in to randomly selected promoter sequences .
a b
a b
a a
aa
aa
aa
a a

Col
-0
Le
r
Bu-
6
Cvi
-0
Go-
2
Fr-
4
In-0
Gr-
3
Sha
Li-
3
Bla
-14
Col
-0
Le
r
Bu-
6
Cvi
-0
Go-
2
Fr-
4
In-0
Gr-
3
Sha
Li-
3
Bla
-14
At4g34135
At3g28740
At2g44130
Fig. S 4
Col
-0C
ol-0
All samples + CJ
RNA loading
RNA loading
Fig. S4. Analysis of cis-jasmone response of twelve different Arabidopsis ecotypes by Northern blot. Four plants per ecotype (4 weeks old) were exposed for 20 hrs to cis-jasmone, harvested, the leaves pooled and RNA extracted. Seven micrograms of total RNA was loaded per lane and the blot was hybridised with the probes as specified, representing the TGA-independent pathway (a) or TGA-dependent pathway (b) as described in Fig. 6b.
TGA-independent
TGA-dependent
a
b