Eukaryotic Translation mRNA tRNA rRNA. -monocistronic -1,000-2,000 bases long -methylated at 5’cap...
-
Upload
griselda-cross -
Category
Documents
-
view
218 -
download
0
Transcript of Eukaryotic Translation mRNA tRNA rRNA. -monocistronic -1,000-2,000 bases long -methylated at 5’cap...
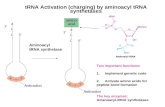
Eukaryotic Translation mRNA tRNA rRNA -monocistronic -1,000-2,000 bases long -methylated at 5cap bases of poly A at 3end -nontranslated 5 reader < 100 bases -nontranslated 3 tailer ~ 1,000 bases - AUG as the INITIATOR -UAA,UAG,&UGA as TERMINATOR mRNA Fig5.16 Expression of mRNA -removal of Poly(A) need for decapping enzyme -Poly(A)-binding protein, PABP on Poly(A) prevents the decapping enzyme from binding to the 5end Life Cycle of Eukaryotic mRNA AU-rich sequence, called ARE ~50 bases in 3 tailer -AUUUA, more repeated -trigger destabilization TRE sequence - contain strm-loop structures for protein binding -Protein inhibit destabilizing tRNA Met-tRNA i for initiation Met-tRNA m for elongation - No formylated Methionine -has an unusual tertiary structure, modofied by phosphorylation of the 2ribose position Eukaryotic Ribosome The right context for Recognization -AUG -the most important seq. GCC A G CCAUGG, purine three bases before AUG and G immediately following it. CAP; for a stable complex to be formed at initiation codon Ternary complex; -Met-tRNA i -eIF2 -GTP Eukaryotic Initiation *form ternary complex *bind free 40S *attach the 5 end Initiation Factors *bind 5 cap *unwind mRNA *bind subunit initiation complex *support 60S joining Elongation Factors: EF-Tu, load aminoacyl tRNA EF-Ts, GDP to GTP Translocation: Peptidyl transferase, synthesis peptide EF-Tu EF-G Fig.6.27