Ernst - Unknown - Comparative Genomics and the Gene Concept
Transcript of Ernst - Unknown - Comparative Genomics and the Gene Concept
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 1/29
COMPARATIVE GENOMICS AND THE GENE CONCEPT
ZACHARY ERNST
A BSTRACT. The gene concept has fallen on hard times in the philosophy of biology. Although
we are confronted on a regular basis with reports that ‘the gene for such-and-such’ has been
discovered, the received view in the philosophy of biology is that current workin genomics shows
that there is no such thing as the gene. In this paper, I argue that such a skeptical conclusion is
unwarranted. In fact, contemporary work in genomics not only shows us that the gene does
exist, but it points the way toward a precise characterization of the gene concept. In the course
of making this argument, I provide an overview of one contemporary approach to gene discovery
and genome annotation that makes crucial use of techniques from computer science.
1. INTRODUCTION
If there is a philosophical consensus on the status of the gene, it would be that current re-
search into molecular biology shows us that the gene is an outmoded concept. John Dupré put
the point succinctly when he said that such modern research was ‘the beginning of the end’ of
the traditional concept of the Mendelian gene. This argument owes much to the work of David
Hull [8,9], whose classic skeptical stance on the reality of the gene has become somewhat of a
received view.
But the received view is mistaken; we have good reason to hold onto a suitably revised gene
concept. In this paper, I will argue that doubts about the gene concept are rooted in a faulty
theory of reference for theoretical terms. When we critically examine how the theory of refer-
ence should be applied to terms such as ‘gene’, then we see that we must attend to the details of
contemporary genomics research if we are to determine whether genes exist. Accordingly, this
paper provides an overview of one approach to comparative genomics research. This research
strongly suggests a revised, but recognizable gene concept. This concept crucially makes re-
course to the evolution of modularity. Thus, while I propose a positive solution to the problem
of characterizing the gene concept, it also turns out that genomics research focuses our atten-
tion on another (and perhaps more important) problem. This is the problem of understanding
why natural selection sometimes seems to favor the evolution of highly modular structures.
Date : April 10, 2008.
Many thanks to Ross Overbeek for his instruction at Argonne National Laboratory, and to Alexander Rosenberg for
saving me from a couple of awful howlers in this paper.
1
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 2/29
2. THE LOGIC OF S KEPTICAL A RGUMENTS
In order to motivate the central argument of this paper, I shall summarize and critique com-
mon arguments that aim to establish that the gene does not exist. When these skeptical argu-
ments have been criticized, we shall have better motivation to examine current research into
genomics in the following sections.
Current research into molecular biology has dashed all hope of a simple molecular imple-
mentation of the Mendelian gene. If we had hoped that genes would supervene on simple,
contiguous, easily identifiable stretches of DNA, then we must at least lower our expectations.
Although classical genetics makes use of notions of dominant or recessive genes, it is now well
understood that such concepts are, at best, useful but severe idealizations. Genes (if they ex-
ist) are neither implemented in a simple, straightforward manner, nor are they inherited in a
simple, straightforward manner.
It is from these uncontroversial premises that skeptics about the gene – including John Dupré
and David Hull – make their arguments. These arguments draw upon premises that are often
pressed into service for anti-reductionist arguments concerning the gene. Indeed, I shall argue
that these arguments are too closely related to these anti-reductionist arguments.
According to these skeptical arguments, the term ‘gene’ is supposed to refer to whatever en-
tity implements the mechanisms of inheritance in a way that approximates classical Mendelian
theory about inheritance. So genes exist only if there is something that does implement inher-
itance in such a way. But when we begin to investigate how various segments of DNA imple-
ment the mechanisms of inheritance, we quickly discover that there is no simple story to be
told. The same, or functionally same, phenotypic characteristics are famously understood to
be multiply realized by many different possible segments of DNA [26]. Furthermore, owing to
complications arising from developmental facts,identical segments of DNA may instantiate dif-
ferent phenotypic characteristics. The point is a familiar one from anti-reductionist arguments,
namely, that the relationship between genotype and phenotype is hopelessly many-many, not
capable of any simple characterization by any finite set of bridge laws.
It should strike us as odd that these premises – which are typically the premises of anti-
reductionist arguments – should be pressed into service to support a non-existence claim about
genes. After all, reductionist theses are typically understood as conclusions about explanations
and terms; that is, reductionism is a linguistic thesis. But existence claims are obviously onto-
logical theses. Alan Garfinkel puts the point succinctly:
So reductionism, which is on its face an ontological question, is really a question
about the possibility of explanation: to say that something reduces to something
else is to say that certain kinds of explanations exist. [5, p. 443]2
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 3/29
Thus, it might appear at first glance to be a non-sequitur when Hull and Dupré argue that genes
do not exist by providing premises of anti-reductionist arguments. So it is important to try to
reconstruct this line of reasoning in more detail.
It is well appreciated that during the Modern Synthesis, it was Mendel’s work on the mech-
anisms of inheritance that allowed Darwin’s theory of evolution by natural selection to be puton a solid theoretical foundation. Although he had no way of guessing as to the physical im-
plementation of inheritance, Mendel’s insight was to recognize that the observed facts of in-
heritance could be explained by positing theoretical entities called ‘genes’ that would somehow
influence the development of organisms, while also following simple rules of transmission from
parent to offspring.
Mendel’s rules of inheritance assumed that these posited entities would fall into various cat-
egories, including ‘dominant’ and ‘recessive’, that each gene would have an equal probability
of being passed along from parent to offspring (the so-called ‘independence of assortment’ as-
sumption), and that they would affect the development of the organism in a straightforwardmanner. Of course, none of these assumptions have been borne out in the long run – inheri-
tance, for example, can be affected by so-called ‘driving genes’, and the mechanisms of assort-
ment are severely affected by the location of particular stretches of DNA along the chromosome.
Specifically, if two stretches of DNA are close together on the chromosome, then the probability
that one will be inherited by the offspring is positively correlated with the inheritance of the
other. So these Mendelian assumptions have turned out to be false.
Skeptics about the gene have used these complications in a deceptively simple argument.
If the theoretical term ‘gene’ refers to an entity that controls inheritance, and which assorts
independently, then there simply is no such thing that answers to that description. Hence, theterm ‘gene’ fails to refer to anything at all; therefore, we are to conclude that genes do not exist.
Hull puts the argument in an interesting way. According to Hull, we have to distinguish two
possible scenarios that could play out in a reduction of one theory to another. On the one hand,
it may turn out that the reduced theory is discovered to be incorrect in some relatively minor
ways; thus, in order to carry out the reduction, we would have to first ‘correct’ it in order to bring
it into line with the reducing theory. Such would presumably be the case when we discover how
to reduce (e.g.) Newton’s law of cooling to statistical mechanics – whereas we originally had
a deterministic and non-probabilistic theory, we ‘correct’ it by introducing statistical factors
into the theory. But according to Hull, this is not a problematic case, because the theory is
recognizably the same both before and after the reduction has been carried out.
On the other hand, it is possible to discover that the reduced theory must be modified beyond
recognition in order to bring it into line with the reducing theory. In such a case, we cannot
simply say that we are ‘correcting’ the reduced theory – instead, we are replacing it. As Hull puts
the point regarding the reduction of classical Mendelian genetics:
3
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 4/29
My intuitive impression continues to be that the differences between the cor-
rected and uncorrected versions of these theories are too numerous and too
fundamental to consider the relationship between the two corrected theories re-
duction in the formal sense of the term. Pre-analytically, the relation between
Mendelian and molecular genetics is a paradigm case of theory reduction, butfrom the point of view of the logical empiricist analysis of theory reduction, it
looks more like replacement. [8, p. 660]
However, the simplicity of that argument belies a deep difficulty concerning the reference of
theoretical terms. For nowhere else do we tie a term’s denotation to its original intensional
meaning. For example, although it is certainly true that the term ‘atom’ was originally intro-
duced to refer to an ‘indivisible thing’, and that there is no such (known) indivisible thing, we
do not conclude that atoms do not exist. Rather, we simply recognize that the original con-ception of the atom was in error. Indeed, if any theoretical term is ‘unrecognizable’ from the
perspective of its original meaning, the term ‘atom’ is.
In general, we feel free to allow the sense of a theoretical term to shift under the influence
of new information concerning that term. Thus, as we discovered that atoms were indeed ca-
pable of being divided into component parts, we simply allowed the term ‘atom’ to continue
to refer to those entities, in spite of the fact that they turned out not to answer to their original
conception. This strategy is underwritten by the causal theory of reference, attributed primarily
to Quine [25] and Kripke [15]. According to the causal theory of reference, proper names and
theoretical terms may initially have their reference fixed with the help of a connotative defini-tion, their reference is in fact fixed by virtue of a causal chain which runs from the user of the
term (e.g. a practicing scientist) back through a series of experiences which may include con-
versation, writing and so on. That chain will eventually terminate in some causal influence that
the entity in question had on someone who fixed the referent of the term by stipulation. The
upshot of the causal theory of reference is that it is this causal relationship, and not a set of
necessary and sufficient conditions, that fixes the referent of a theoretical term. In this way, we
are able to account for the continuity of a scientific theory in the face of radical theory change.
For although the meaning of a theoretical term may eventually change to the point at which it
is unrecognizable to its original users, the causal chain leading from that entity to the users of
the term remains.
For the present purposes, the lesson is straightforward. We do not attempt to defend the
view that the Mendelian concept of the gene is alive and well. But we ought to question Hull’s
assumption that there is any cut-off point after which the term has been so dramatically revised
that it loses its ability to refer to the same entity. We should not expect the meaning of the term
‘gene’ to remain fixed in the light of ongoing scientific research any more than we should expect4
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 5/29
the term ‘atom’ to retain its original connotation. It is a mistake to assert that any set of con-
ditions can be attached to the term that are necessary for that term to refer. Specifically, con-
tiguity of the chromosome, independence of assortment, a simple developmental story from
genotype to phenotype, and all such other conditions are not necessary (singly or jointly) for
the term ‘gene’ to refer.
2.1. Indispensability. According to a line of argument that has become widely accepted, we
are justified in claiming that a theoretical term refers to a real entity if the use of that term is
indispensable in explaining observed phenomena. Normally, however, when we are able to for-
mulate bridge laws relating some supervenient entity A to its underlying physical implemen-
tation B in a straightforward, suitably non-disjunctive way, then that reduction may be taken
to show that we can replace any mention of A in our explanations with a translation into thelanguage of B . In other words, when we have a successful reduction in hand, that is taken to
show that the reduced entity is dispensable . Thus, if a reduction is evidence at all concerning
existence, then it should speak against the existence of the reduced entity. Conversely, when
we find that we are unable to carry out a reduction, then we will typically assume that we are
correspondingly unable to eliminate the term in question. Thus, the use of that term is more
likely to be in dispensable.
For example, suppose that a metaphysical argument is proposed that only basic substances
such as subatomic particles exist, but not the ordinary objects such as tables and chairs that we
ordinarily take to be composed of those basic substances. Such metaphysical arguments typi-cally proceed by showing that there is no explanation or causal power possessed by tables and
chairs that cannot be fully explained by the causal powers of the particles that (we ordinarily
take to) compose tables and chairs. Thus, the argument goes, we can – at least in principle – re-
place any talk of these ordinary objects with talk of basic substances. And so, the dispensability
of these entities is taken as defeating any reason to believe that they do exist.
So regarding the gene, we find that there is a tension between antireductionist arguments
and arguments purportedly establishing that genes do not exist. For normally, the premises
of antireductionist arguments are taken to imply that the unreducible entity is in dispensable,
and that we therefore have reason to believe that the entity exists. On the other hand, if the
entity in question can be reduced, then the use of that term is dispensable , and we thereby
lack at least some important justification for saying that the entity exists. But if we were to
accept the arguments of Hull and others, then the gene is completely different. For they take the
premises of antireductionist arguments to show that genes do not exist. This tension between
antireductionism and indispensibility provides a further reason to question such arguments
purporting to show that the gene does not exist.5
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 6/29
3. FUNCTIONAL CHARACTERIZATION OF GENES?
In the best of all possible worlds, we could simply define each particular gene as a particular
sequence of nucleic acids, located in a specific place on the chromosome. It is a point not worth
belaboring here that such a definition is hopeless [26]. Obviously, if genes do exist, then there
will be numerous small changes to the particular sequence of nucleic acids that will not affect
the identity of the gene. Furthermore, as philosophers of biology have long understood, the
same gene type may be tokened at two or more different locations on the chromosome without
affecting the identity of the gene. Indeed, such shifts appear to play a crucial role in evolutionary
processes, and the reconstruction of the history of such changes gives us valuable insight into
the evolution of various species.1
For a philosopher of science, when a physical characterization fails, the obvious next step is
to try for a functional characterization. That is, for any particular gene, we may try to define it
using the following schema:
(3.1) Gene X =def any nucleic acid sequence performing function F
Unfortunately, as is also well-appreciated by philosophers of biology, it is common for a par-
ticular sequence that performs one function in some species to perform a different function in
another species. Intuitively, we would like to be able to claim – if we were to have a workable
gene concept – that the same gene performs two different functions. However, schema (3.1) will
not countenance such a claim. Of course, one could always hold out for a disjunctive version
of schema (3.1), but there is no a priori way to set an upper limit on the number of possible
functions that a gene could perform. One could reasonably suspect, in fact, that without set-
ting an arbitrary limit on the number of possible contexts in which a particular sequence might
appear, that there is no upper limit to be had at all. Thus, it looks as if neither a physical and
reductive definition, nor a functional non-reductive definition will work for defining the gene.
No wonder, then, that philosophers of biology have despaired of coming up with a workable
definition of the gene.
4. C AUS ATIO N AND THEORETICAL TERMS
At this point, we have a trio of problematic proposals regarding the reference of the theoret-
ical term ‘gene’. First, we have the traditional Mendelian gene concept, which is well-knownto be incorrect, or at least to be so severely idealized that it is not to be found in the genome.
Second, we have the philosophical positions advocated by Hull and Dupré, according to which
the term ‘gene’ simply fails to refer at all. But as I have argued above, their negative arguments
ultimately fail because they rely upon problematic theories about the reference of theoretical
terms. Third, we have the possibility that a functional characterization of the gene concept can
1See below, in section 6.
6
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 7/29
be made out. But for familiar reasons having to do with multiple realizability, this approach
fails as well.
These difficulties may properly be considered symptoms of a deeper problem regarding the
gene concept. For the question of whether the gene exists should be interpreted as the question
of whether the theoretical term ‘gene’ successfully refers. Thus, the question of whether thegene exists is primarily a question for the philosophy of language and specifically for the theory
of reference. And the gene concept provides a particularly difficult test case for a theory of
reference.
As I have argued above, it is too quick to argue that the term ‘gene’ fails to refer merely be-
cause our current understanding of genetics demonstrates that the Mendelian gene concept is
inadequate. For such an argument implicitly depends upon a theory of reference that fixes that
reference of a term by giving something like a definite description of it. And such a picture has
been long recognized to be inadequate for the task of accounting for theory change. Thus, we
should not be surprised to find that such a theory of reference turns out to be inadequate forcharacterizing as complex a theory as that of genetic inheritance. Accordingly, a defense of the
gene concept requires (at least an outline of) a defense of a theory of reference that is plausible
on its own, while remaining compatible with the view that the term ‘gene’ successfully refers.
Unfortunately, the subject of the reference of theoretical terms is far too complex for the cur-
rent paper. However, I think that it is possible to argue that a causal theory of reference allows
us to retain a meaningful gene concept. That gene concept is one that emerges as a result of
current research into genomics. Furthermore, standard objections to the causal theory of ref-
erence – as it is applied to theoretical terms – are problematic. This will be the subject of the
current section.
4.1. Ostension and Theoretical Terms. The obvious alternative to a theory of reference that is
based on definite descriptions or other intensional meanings is a causal theory. Indeed, the
causal theory of reference has become the received view for theoretical terms precisely because
it is capable of accounting for how terms maintain their reference while their sense changes
significantly. Thus, adopting a causal theory is a promising strategy for accounting for the gene
concept.
However, we immediately run into difficulties if we try to straightforwardly apply the causal
theory to this case. For on a standard picture of the causal theory, a term acquires its reference
through an initial ‘baptism’, in which a demonstrative is used to fix the reference of a term. For
example, a parent may fix the reference of the term ‘Joe’ by indicating a child and using the
demonstrative, ‘that child shall be called ‘Joe’ from now on’. Thus, the reference of a name may
be fixed without having in mind a definite description of the object named. Furthermore, when
a person uses the name to refer to the object, she may successfully do so despite the fact that
her own understanding of the object’s properties are quite incorrect. So long as their use of the7
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 8/29
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 9/29
are what we might call ‘hybrid theories’, which are causal, but require intensional information
about at least some of the terms in order to fix their reference – the theories of Enç and Nola are
examples of this kind of hybrid theory.
The reference of the term ‘gene’ is threatened under any hybrid theory of reference, since the
intension of the term has obviously changed a great deal in the history of genetics. However,hybrid theories of reference face difficulties because the distinction between ostensible and
non-ostensible terms is extremely problematic. This is simply because the ability of an entity to
be directly observed just is a particular kind of causal power the entity possesses. Thus, even an
entity that is directly ostensible is ostensible because it has the causal power to affect our sense
organs in a particular way. This is apparent in Kripke’s discussion of the term ‘heat’, where he
describes the causal powers of molecular motion in terms of their ability to create certain effects
in our nervous system. In characterizing the manner in which we ostensibly refer to heat,Kripke
seems to equate reference by direct ostension with reference by more indirect methods:
At any rate, we are able to identify heat, and be able to sense it by the fact that that
it produces in us a sensation of heat. It might here be so important to the concept
that its reference is fixed in this way, that if someone else detects heat by some
sort of instrument, but is unable to feel it, we might want to say, if we like, that
the concept of heat is not the same even though the referent is the same. [15, p.
131]
In short, because the ability to be observed is an instance of a causal power that could figure
into the use of the schema (S), it is far from clear how to draw a distinction between ostensible
and non-ostensible terms.
2
But even if we put aside this difficulty for the time being, we can stillidentify two two general types of cases that have traditionally been used to motivate a hybrid
account of how the reference of theoretical terms is fixed. These two cases are:
(1) cases in which the intensional meaning of a term is inadequate for fixing its reference,
and
(2) cases in which we are more likely to abandon the term rather than radically revise its
intensional meaning.
My contention here is that by attending to these cases, we are led to a better modification of the
theory of reference for theoretical terms, and that this modification makes sense of the contin-
ued use of the term ‘gene’. I shall discuss each in turn, before outlining the positive proposal.
2The difficulty of distinguishing between ostensible and non-ostensible terms is parallel to the familiar difficulty
of distinguishing between observable and non-observable entities. For ‘direct’ observation requires the observed
thing to exert a causal influence upon our sense organs and a (perhaps implicit)theory of how theresulting sensory
impressions reveal facts about it. In fact, I think it is reasonable to suspect that the distinctions between ostensible
and non-ostensible entities on the one hand, and observable and unobservable entities on the other, stand or fall
together.
9
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 10/29
4.2. Thales and the amber. Nola has contended that if the bare causal theory were correct,
then people would be in a position to fix the reference of theoretical terms when they clearly
lack the necessary level of understanding to do so. In particular, anyone who was in a position
to observe the effects of some theoretical entity would be able to stipulate a name for whatever
it is that happens to be the cause of those effects. But according to Nola, it is clear that (at leastin many cases) more is required.
For example, Nola recounts a story about Thales, who observed (what turned out to be) the
buildup of electrical charge on a piece of amber after it had been rubbed. If the bare causal the-
ory were correct, then Thales would have been in a position – with no further information about
electricity – to stipulate a term for ‘whatever it is that causes the attractive effects of amber after
it has been rubbed’, and would thereby have fixed the reference of a term upon electricity. But
according to Nola, this sort of case should strike us as wrong – it attributes ‘too much scien-
tific prescience to Thales in the absence of any theory about the item so picked out’ [18, p. 516].
Rather, in order for Thales to have successfully picked out electricity, he would have had to havehad some theory about how the entity causally brings about its effects.
However, even if we share Nola’s intuitions about Thales’s alleged inability to fix the reference
of any term upon electricity, there is still a difficult problem with requiring that Thales would
have to have had a theory about how electricity causes the attractive powers of the amber. This
difficulty can be brought out as a dilemma, for we must either require that the theory be correct
(or nearly correct), or we must waive the requirement. It should be clear that the first horn of
the dilemma is unattractive for two reasons. First, it is plainly too demanding, and would put
the cart before the horse in that it often turns out that it is necessary to fix the reference of a
term before engaging in the kind of research that could lead to the correct theory about theentity’s causal powers. Second, if we require a correct theory of the entity’s causal powers, then
we are treading too closely to a definite description theory of reference – for the correct theory
could simply be used to fix the reference of the theoretical term without having to worry about
a causal theory of reference at all.
But we cannot weaken the requirement of truth, either. For suppose that we require that in
order for Thales to be able to fix the reference of the term, he need only have some theory or
other – even a false one. Although it is certainly true that a term may have its reference fixed
in spite of the fact that the intensional meaning of the term is wrong, it is strange to require
such a theory, while admitting that it might be totally false. To put the point rhetorically, it is
fair to wonder what a false theory adds to the reference-fixing ability of Thales that cannot be
otherwise be met while being agnostic about how the entity causes its observable effects. I thus
conclude that cases such as this one do not pose a difficulty for a bare causal theory of reference.
4.3. Phlogiston. We need now to consider cases in which the use of some term is abandoned
as we discover new information suggesting that the term fails to refer. The standard example10
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 11/29
of this phenomenon is the failure of the theoretical term ‘phlogiston’ to refer to any real entity.
Enç and Nola both argue that the reference of ‘phlogiston’ was to have been fixed partially in
virtue of the intensional meaning of the term; thus, when it was discovered that its intensional
meaning was not satisfied by any real entity, that discovery ‘was tantamount to discovering that
phlogiston did not exist’ [2, p. 271].But the interesting feature of this example, which makes it not good support for any hybrid
theory of reference, is that the intensional meaning of the term was inextricably bound up with
the causal powers that were attributed to phlogiston. The following discussion from Enç is in-
structive:
For example, in the phlogiston case, when the term “phlogiston” was introduced,
it was at least believed that whatever causes fire can saturate air during combus-
tion and that when the air is saturated the fire dies out... Furthermore, the belief
that this substance had the power to restore the metallic properties of calx and
to lead to death by suffocation... led to the belief that the substance in question
was a new kind of substance. [2, p. 271].
Thus, when these beliefs were discovered to be false – i.e. that there is no substance meeting
that description – scientists concluded that phlogiston does not exist. From this, Enç concludes
that ‘in introducing a term, the scientist is not just naming whatever it is that is responsible for
such and such phenomena, he is rather naming a kind of object partially specified by the kind-
constituting properties he believes the object to have and by the context in which the object
plays its explanatory role’ [2, p. 271]. According to this argument, a bare causal theory would
have it that the scientists were referring to oxygen (since oxygen is what is responsible for com-bustion), and they would merely have discovered that ‘phlogiston’ actually refers to oxygen, but
that some of their other beliefs about phlogiston were false (for example, that it is responsible
for suffocation).
Kyle Stanford and Philip Kitcher call this the ‘no failure of reference problem’ for the causal
theory [28]. In general form, the problem is that so long as the person who introduces the term
defines it as ‘the cause of X ’, where X is some real effect of some cause or other, then the term
is guaranteed to refer to that cause, whatever it may turn out to be. But their intuition, which is
plausible enough, is that if the cause turns out to be totally different from what the introducer
of the term has supposed it to be, then we are better off judging that the term fails to refer at all.
However, it is not so clear that the bare causal theory of reference really does lack the re-
sources to yield the correct judgment that ‘phlogiston’ fails to refer. In short, I think it is fair
to say that those who use this particular episode in the history of science have cherry-picked
certain features of the example. To see this, consider a simplified and fictional case resembling
the historical example. Let us suppose that a scientist we shall call ‘Williams1’ inquires as to the
cause of combustion, supposing that there may be some such substance, and he accordingly 11
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 12/29
defines ‘phlogiston1’ as ‘whatever substance causes combustion’. Our fictional scientist may
develop all sorts of other beliefs about phlogiston1, many or all of which may be mistaken. He
may believe, for example, that it is emitted from burning bodies or that it has a negative weight.
But let us suppose that his original reference-fixing stipulation makes recourses only to the
particular causal property of causing combustion. When Levoisier discovers oxygen, Williams1may quite reasonably assert that ‘phlogiston1 is oxygen’, in spite of the fact that many of his spe-
cific beliefs about phlogiston1 will have to be revised or abandoned completely. And of course,
this is just what the bare causal theory would have be the case.
Now let us complicate the example somewhat. Suppose that another scientist – Williams2 –
inquires as to the causes of combustion and suffocation, hypothesizing that some substance is
the common cause of both. Then he stipulates that ‘phlogiston 2’ shall refer to ‘whatever sub-
stance is the cause of combustion and suffocation’. Like his counterpart, he may form a variety
of other beliefs about this new substance, but these play no role in fixing the reference of the
term ‘phlogiston2’. Also like his counterpart, Williams2 stipulates the reference of ‘phlogiston2’according to schema (S) above, but in this case,Φ is conjunctive.
According to the ‘no failure of reference’ objection, the bare causal theorist is committed to
the untenable thesis that ‘phlogiston2’ refers to something , when in fact, it fails to refer to any-
thing at all. However, the bare causal theory has the resources to yield the correct conclusion.
After all, there is no substance whatsoever that is both the cause of combustion and suffocation.
So the bare causal theory does not erroneously say that ‘phlogiston 2’ refers to oxygen (or any
other substance). Rather, a bare causal theory may rightly conclude that the term simply fails
to refer at all.
This example suggests that when the reference of a theoretical term ‘T ’ is fixed by stipulating that refers to whatever is the cause of Φ, then it is possible that ‘T ’ will not refer if there is no
single kind of entity that is the cause of Φ. One way that this can happen is if it is supposed that
Φ has some singular cause, but in fact, two or more different kinds of entity are the cause of
Φ. And of course, this is precisely the type of case which proponents of hybrid theories use for
support.3
A variety of other objections have been made to the bare causal theory of reference.4 I believe
that these other objections can be met. However, because the purpose of this paper is simply
to defend the reference of one particular theoretical term – ‘gene’ – I shall assume at this point
that I have sufficiently motivated some doubts about the need for adopting a hybrid theory of
reference.
3To take another example, Enç [2] discusses a hypothetical case in which Jones uses the name ‘Snowwhite’ to pick
out the entity – whatever it is – that ate his lettuce and carrots last night. As the example proceeds, however, Jones
attributes many other events to Snowwhite (e.g. breaking Jones’s teacup, getting into the peanut butter). And as
these other causal powers are attributed to Snowwhite, Enç motivates the intuition that Jones is not successfully
referring to anything. But this case may be dealt with in the same way as the phlogiston example above.4For instance, see Kitcher’s discussion of the so-called ‘qua problem’.
12
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 13/29
5. CONNECTING REFERENCE TO RESEARCH
I have argued that standard objections to the bare causal theory of reference ultimately fail,
and that in particular, criticisms fail that have been leveled against the term ‘gene’. However,
there is a motivation of these criticisms that is worth examining in more detail, with the aim
of saying something more positive about the reference of theoretical terms. The motivation for
criticisms of the causal theory seems to be that when the intensional meaning of the term has
changed beyond recognition, the research program within which the term was to play a role
must be abandoned or changed entirely. Frederick Kroon expresses this motivation explicitly:
Once again, then, the burden of reference for the term introduced rests broadly
on the theory within which the term is embedded, and not on some cautious
causal descriptions of the form: ‘whatever it is that is responsible for such and
such phenomena’... [16, p. 50]
Here, I think that Kroon uses a correct observation to support a criticism that is too general.Specifically, Kroon is right to say that ‘the burden of reference... rests broadly on the theory
within which the term is embedded’. But when Kroon goes on to say that the ‘cautious causal
description’ – what we have been calling scheme (S ) – does not underpin the reference of the
theoretical term, this suggests the dubious position that the causal description of the entity can
be separated from the role of the referring term in the underlying research programme.
However, the nature of the research programme within which the term is embedded is de-
termined largely by the causal powers we attribute to the referent of that term. For example,
Kroon considers the case of Neptune, which was used by Kripke to support his causal theory
of reference. Kroon asks us to consider the following purported counterexample to the causaltheory. Suppose that the term ‘Neptune’ was introduced to refer to whatever it is that is the
cause of some observed perturbations in the orbits of various planets. Of course, the original
intensional meaning of ‘Neptune’ was include the proposition that the entity is an unobserved
planet. Now suppose we were to discover that, through a very indirect and subtle route, Earth
is responsible for the observed perturbations.
According to Kroon, the causal theory of reference is committed to the view that ‘Neptune’
refers to the planet Earth, whereas the correct conclusion is that the term ‘Neptune’ does not
refer at all. Although this may be the correct conclusion to draw in this specific case, I think
that Kroon, Enç, and Nola have misdiagnosed the motivation for abandoning theoretical terms
(when it is appropriate to do so). For what motivates us to abandon a particular theoretical term
is that the entire research program ‘within which the term is embedded’ is given up. In contrast,
Kroon, Enç, and Nola assume that the divergence from some intensional meaning of the term
is what is responsible for the failure of the term to refer. But these are distinct phenomena – it
is possible for the intensional meaning to change without dramatically affecting the research
programme, and vice-versa.13
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 14/29
To see this, let us consider variants on Kripke’s Neptune example. Suppose scientists stipu-
late that the term ‘Neptune’ shall refer to whatever (heretofore unobserved) planet causes the
observed perturbations in the orbits of other planets. Here, our research programme into the
nature of Neptune would be to calculate – based on Newtonian physics – what the mass and
position of such a planet would have to be. Then we would try to observe whether there was infact a new planet at the expected location.
Now consider two different ways in which such a research programme could yield surprising
results. First, suppose that after the appropriate calculations, it turned out that there was not
a planet, but a large asteroid or other body in the appropriate location. In such a case, the
intensional meaning of the term ‘Neptune’ would have to be revised dramatically. However,
we would not conclude that Neptune does not exist; instead, we would conclude that the term
‘Neptune’ has turned out – surprisingly – to refer to an asteroid instead of a planet.
In contrast, consider a scenario like the one discussed by Nola. In this second case, it turns
out that our understanding of gravitational attraction is dramatically wrong; it is not an unob-served planet that causes the perturbations, but the Earth (through a circuitous and surprising
route). In this case, Nola is right when he says that the correct conclusion would be that the
term ‘Neptune’ does not refer at all.
In both cases, the intensional meaning of the term is importantly wrong; in the first case, it
turns out that there is no planet that causes the orbital perturbations. In the second, it turns
out that there is a planet, but not an unobserved one. Note that it would be a mistake to con-
clude that the intensional meaning was obviously more mistaken in one case than in the other.
For in one case, Neptune turns out not to be a planet at all; but in the second case, there is at
least a planet (namely, Earth) causing the observed phenomena. So if the cases yield differentintuitions about the reference of the term ‘Neptune’, it is not because of obvious differences
regarding their intensional meanings. Rather, what explains the difference between these two
cases is that the research programme for investigating the cause of the observed phenomena
must be given up entirely in the second case, while it remains intact in the first case. Accord-
ingly, we judge that the term continues to refer when the research programme remains intact;
but when the research programme must be given up, we judge that the term fails to refer.
Although it is obviously a difficult question as to when a particular research programme has
been given up, even a rough-and-ready judgment is good enough to make sense of traditional
examples that are used in discussions of the reference of theoretical terms. But more impor-
tantly, we better understand why a bare causal theory of reference is so plausible when it is
applied to theoretical terms, and why it seems to fail in some especially problematic cases.
Clearly, when one baptizes a theoretical term via schema (S ), and thereby attributes some
causal power to the putative entity named by the term, then that attribution will guide research
into the nature of that entity. For example, if one supposes that phlogiston or oxygen is the
14
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 15/29
cause of combustion, then a researcher will try to discover the nature of phlogiston or oxygen
by observing what happens when combustion occurs. If one were to discover – as in the case of
phlogiston – that the causal powers of the entity were radically misdescribed (perhaps because
nothing really has those causal powers) – then the research program must end or be revised
beyond recognition. In such a case, the natural conclusion to draw is that there is no referent of the problematic theoretical term.
This suggests that the plausibility of the causal theory for theoretical terms stems from the
relation between the causal powers of an entity and the relevant research programme. One
may explain why a theoretical term refers or fails to refer by citing the appropriate facts about
the research programme, not by citing facts about the original intensional meaning of the term.
As the research programme evolves, the intentional meaning may change without losing the
referent of the term.
If my arguments so far are sound, then the lesson for reconstructing the gene concept is
straightforward. We must place primary importance on understanding the research programmethat purports to discover genes and elucidate their properties. If we want to understand whether
the term ‘gene’ refers, then we must understand whether contemporary research into genes
is actually tracing observed phenomena back to a referent of the term ‘gene’. The question of
whether this is indeed occurring may be determined only by understandingthe methodological
assumptions that are required by contemporary research. Thus, a discussion of contemporary
genomics is required.
6. COMPARATIVE GENOMICS – A BIASED O VERVIEW
For the philosophy of biology, genomics provides an extremely valuable area of research. The
novelty of this methodology and the startling successes of genomics raise philosophical issues
that deserve a great deal of attention from philosophers of science. Furthermore, in addition to
raising new problems for study, the field of genomics also helps us to settle existing problems
relating to the definition and reference of theoretical terms, the status of reductionism, the role
of information processing technologies in the special sciences, and a host of other issues.5
However, the techniques used in genomics are so unfamiliar that it does require some time
to become sufficiently acquainted with them. So in this section, I shall offer a biased overview
of one current approach that is making fast progress toward identifying genes, and determin-
ing the function of particular genes. This is merely one such approach – no representation is
made here that it is the best approach (on any particular measure). But I do allege that it is an
extremely informative approach, deserving of careful study by philosophers of biology.
In what follows, I shall use the term ‘gene’ uncritically, following the usage that has become
standard in genomics research. In later sections, I shall turn to a critical analysis of this concept,
5Some of these other issues raised by contemporary genomics are surveyed in [3].
15
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 16/29
and I shall argue that a useful and fairly traditional gene concept can be elaborated from this
usage.
6.1. Preliminaries. What makes it possible for an outsider to understand this particular re-
search programme is that the methodology outlined here is highly abstract – so abstract, in fact,
that many biological details may be omitted. So here, I shall give an overview of this research at
a high level of abstraction.6
First of all, it is useful to distinguish between two complementary projects in genomics re-
search. The project that is most familiar to philosophers of science as well as to the general
public is genome sequencing – this is the process of making a catalogue of the specific sequence
of nucleic acids that comprise the genome of a particular species. After this process is com-
pleted, we are left with an immensely long sequence of the familiar A , T ,G ,C characters that
standardly represent the genome. Of course, the most famous gene sequencing project is the
human genome project, which has successfully completed the sequencing of an entire human
genome.
However, for our purposes, the more interesting project is genome annotation . In many ways,
this is the more difficult project – for it aims at extracting useful information from the nucleic
acid sequences that are provided by genome sequencing. Genome annotation includes the
process of so-called ‘gene discovery’, as well as the extraction of information about how the
genes function together to implement the processes that are required for the organism. It is
the difference between genome sequencing and genome annotation that explains why signifi-
cant advances in gene therapy, diagnosis, and other areas did not follow immediately upon the
heels of the human genome project. For those advances require genome annotation, for which
genome sequencing is merely a necessary preliminary step.
6.2. The Subsystems Approach. Much of the research currently being conducted in genomics
concerns the synthesis of various compounds that are required for the cell to function. Particu-
larly, the process of synthesizing these compounds consists of absorbing nutrition through the
cell wall and driving it through a multi-stage process in which various intermediary compounds
are gradually transformed into others, eventually resulting in the final synthesis of the required
chemical.
At this point, we must introduce the necessary vocabulary for describing such a process at a
sufficiently high level of abstraction.7 We shall use the term subsystem to refer to any multi-stageprocess that takes as input a particular chemical compound and outputs a new compound that
is synthesized by the cell. These subsystems may be multiply-realized – that is, there may be
6Indeed, it is an interesting feature of genomics research that it is common for computer scientists with no formal
training in biology to play an important role. This is both due to, and the cause of, the high level of abstraction that
is so common in genomics research.7Here, I outline themethodology and employ theterminology used in a series of papers primarily by Ross Overbeek
and Rick Stevens [19–23].
16
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 17/29
!!"#$%&'!()!*+,-.!/!.0.1,+$'*%$
234!!!*#$%&'+56*+.&'!
(!*'*-&'!3!7'5%*+&'!+$8.!
*'*-&'!3!7'5%*+&'!+$8.!)9/!
:,*+,-.1,+$'.&'!3!
*'*-&'!3!*'*-,-$
234!!!*#$%&'+56*+.&'!(!
);<;=;>? );/;=;!
<;@;=;AB
:,*+,-.1,+$'*%$
!!"#$%&'!((!)9/!
/;<;);=</;<;);=C
/;=;=;/
!!D5##,-&'!
(!)!*+,-.!
/!.0.1,+$'*%$
(!D5##,-&'!((!)9/
)9/!:,*+,-.1,+$'*%$
(!)9<!3,E&:6.:,1,#.',-*%$(!A!"81*6%&' 1E.81E*%$
(!"81*6%*%$ A!8$+,*':$E&:$
(!G.+.8$6,-$
);B;);A =;);=;==
=;=;=;<
A;);=;@) =;<;=;)/ );/;=;=B
<;@;=;=>
@;=;=;B
A;=;=;)C
);<;=;==B :,*+,-.1,+$'*%$
(!(&8,-$
(!)9<9A9@!
H$%6*E&:6.!
:,1,#.',-*%$
((!)9/!3,*+,-.1,+$'*%$
+$8.!)9/!3,*+,-.1,+$'*%$
%I!"
J'&#,-$
85K8&8%$+K,.8&-%E$8,8"'L*'.,:
+$%*K.',8+
(!(&8
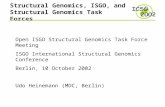
FIGURE 6.1 . Subsystem diagram for Lysine biosynthesis.
many different combinations of distinct steps that will transform the same input compound
into the same output compound. Each of these possible implementations shall be referred to
as a pathway . So in the language that is usually associated with antireductionist arguments in
the philosophy of biology, we say that the same subsystem may be multiply realized by many
different pathways.
We may thus represent any particular pathway by a diagram that resembles a directed graph;
each vertex of the graph represents a discrete step in the pathway, where that step is responsi-
ble for performing one transformation of a chemical compound into a different chemical com-
pound (and possibly giving off a different compound as a by-product). Genomicists refer tothese discrete steps as functional roles . Thus, a pathway is said to consist of a discrete ordered
set of functional roles.
The various possible implementations of a subsystem may be represented simultaneously
in one diagram, which we shall call a subsystem diagram . This is like a graph of a pathway,
except that it is the union of the set of possible pathway implementations. Thus, a subsystem
diagram will typically have branches representing thedifferent paths and sets of functional roles
by which an input compound may be transformed into the required output.
Genes are taken to be sequences of nucleic acids on the chromosome that synthesize the
proteins implementing a particular functional role. So genomics researchers assume that for
any particular functional role appearing in a pathway, there will be a corresponding gene im-
plementing that role. As I shall argue later, this quick gloss is not the full picture of what a gene
is, but it is the preliminary, rough-and-ready notion that is used in genomics research.
With this hierarchy in mind – consisting of subsystems, pathways, functional roles, and genes
– we can describe the major problems of genome annotation that are most important for ge-
nomics research. Because almost every living organism will have to perform many of the same17
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 18/29
tasks at the cellular level, subsystems frequently reappear across many different species. For ex-
ample, one compound – biohistidine – must be synthesized by virtually any living thing. Thus,
some token of the biohistidine synthesis subsystem will have to appear in the central machin-
ery of the cell in almost every living organism. However, multiple realizability ensures that this
subsystem may be implemented by more than one pathway, with potentially many differentgenes implementing the necessary combination of functional roles.
We have an open problem of genome annotation when we discover that some species must
implement (e.g.) the biohistidine synthesis subsystem, but we do not know either which path-
way is the appropriate token of that subsystem, or which genes implement the functional roles
of the pathway. This sort of problem has been called the ‘missing genes problem’, and the pro-
cess of discovering the genes that implement those functional roles is one of the most interest-
ing activities from a philosophy of biology perspective, for reasons that will become apparent.
6.3. Evidence Available Through Genomics Research. An important advantage to the frame- work outlined above is that any given missing genes problem can concisely be represented in a
simple spreadsheet diagram. Indeed, the perspicuity of this representation of the missing genes
problem is an important clue to the right gene concept, or so I shall argue below.
The comparative genomics approach to the missing genes problem takes advantage of the
fact that many nucleic acid sequences are orthologs, where an orthologous sequence is one
that performs a related function in two or more species, and whose appearance in the genomes
of those species is due to common descent (thus, orthologs are a particular type of homologous
trait – see Sober [27]). Thus, partially-completed genome annotations from other species may
provide important clues for solving missing genes problems that arise for other species.Once a missing genes problem has been specified, the genomics approach to its solution
begins by constructing a spreadsheet. That is, we create an inventory of the known implemen-
tations of the subsystems in question – this information may be accessed through public and
private databases, including the KEGG map database8, which I rely upon throughout this sec-
tion. Particular attention is paid to available genome data from species that are known to be
closely related to the species in question because they are more likely to contain orthologous
sequences.
After a set of species has been identified with known or partially known implementations of
the subsystem, that information is organized into a spreadsheet. This representation clearly
highlights the exact information that is available for the target genome, and the information
that is missing. When the spreadsheet has been compiled, it is easy to see how the various
annotated genomes for other species implement the subsystem. In particular, the spreadsheet
representation makes it clear how various functional roles cluster together; it shows how the
8This database me be found at http://www.genome.jp/kegg/pathway.html.
18
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 19/29
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 20/29
implementation of a pathway, then this information may be used to confirm the hypothesis
that another organism uses the same sequence in the same way, provided that the two species
are appropriately related.
Of course, confirmation relations are typically symmetric – if one piece of evidence confirms
another, then the reverse is also true. Thus, there is a feedback between inferring phylogeny andannotating the genome. When we better understand how genomes are annotated, this informa-
tion provides important clues about the evolutionary history of the species and its relationship
to other species. Indeed, the core machinery of the cell evolved so long ago (in comparison to
other traits that are less central to the operation of the organism) that genome annotation of
those subsystems allows us to look further back in evolutionary history than a similar analysis
of other phenotypic traits would allow.9
6.4. A Simple Example. Figure (6.1) is adapted from the KEGG pathway database – a freely-
accessible database of information about known pathways and subsystems in many different
species. It shows the subsystem that synthesizes lysine. One may think of the diagram as repre-
senting all the known pathways by which lysine is synthesized from other chemicals. Boxes in
the diagram with a period-delimited set of numbers – called the ‘EC number’ – represent func-
tional roles, and the circles represent the chemical product that is produced after that func-
tional role has operated. The arrows are used to show the order of steps by which the functional
roles produce the various compound that are necessary for the synthesis of lysine.
As is typically the case, the product of this subsystem may be used by other subsystems to
produce other compounds that are required by the cell. Accordingly, the subsystem diagram
indicates that lysine may be used as an input to the alkaloid biosynthesis subsystem, and thatL-Homoserine may be used in the glycine metabolism subsystem. As we have seen above, any
given subsystem may be implemented by one of several different pathways. These options are
shown in the subsystem diagram by places where there is more than one arrow leading from a
circle.
Figure (6.2) is a representative spreadsheet diagram for a portion of the lysine biosynthesis
subsystem. It collects a portion of the available information for nine bacterial genomes; this in-
formation is taken from the current version of the KEGG database. It corresponds to a portion
of the subsystem diagram (6.1). In the spreadsheet, the species names are listed along the left
side; the various functional roles (indicated by their EC numbers) are listed at the top. A dark-ened rectangle means that the species has an identified sequence implementing the functional
role. Where the rectangle is empty, there is no known implementation of that functional role.
In the spreadsheet, I have divided the functional roles into two groups, which are labeled (A)
and (B). If we examine the functional roles from each of these two groups, we see that there is
9Indeed, one of the reasons for focusing on the central machinery of the cell is that some researchers hope that by
so doing, it will finally become possible to make reasonable hypotheses about prebiotic evolution.
20
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 21/29
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 22/29
implementation. By the same token, we might conjecture that Lysteria monocytogenes imple-
ments role 3.5.1.47.
Clearly, the quick generation of such conjectures is a highly valuable feature of the compara-
tive genomics approach. Furthermore, due to the presence of orthologous sequences, we have
reasonable – but defeasible – hypotheses about how those functional roles are implemented.Specifically, we should look at those sequences that are known to implement those functional
roles in other species. For example, if we wonder which gene implements role 2.3.1.89 in Strep-
tococcus pneumoniae , then it is a reasonable first assay to examine the genome for sequences
that are similar to the ones implementing that role in other bacterial species such as Listeria
monocytogenes , Staphylococcus aureus , and Bacillus subtilis . It is now very simple to conduct
such a search in an automated fashion, since the genome data is simply digital information that
can be searched like any other large dataset.
This example should make it clear why researchers are optimistic about the progress that is
possible in genome annotation. For although this is a simple example, it does faithfully show that there are three distinct stages of genomics research. We may think of those stages roughly
in the following way.
• Formulation of the problem . A missing genes problem can be formulated by discovering
which functional roles appear to be missing from the annotations of species. This can be
automated by considering subsystem diagrams as directed graphs, and then identifying
which paths through the graph are only partially annotated.
• Search through reference genomes . Other genomes can be identified that are known to
implement the missing functional roles. Those sequences serve as models for candidate
sequences in the target genome.
• Confirmation of the hypothesis . If such a sequence is discovered in the target genome,
we may obtain confirming evidence by testing whether the sequence is clustered on the
genome with other sequences that are required for the pathway.
Of course, it may turn out that no such sequence is discovered in any of the comparison genomes.
But that would not show that the comparative genomics approach fails in that case. For if
there is some unknown sequence that implements the functional role in that particular species,
then it is quite reasonable to suspect that the same sequence will implement that role in other
species. So this suggests that a search through other genomes that are also lacking an identified
implementation of the functional role. If there is a sequence that is nearby on the chromo-
some, and which is found in several of the target genomes, then one may conjecture that it
implements the functional role. This is an important point, because a comparative genomics
approach is not limited to cases in which the sequence has already been discovered in some
other species by traditional ‘wet lab’ techniques. Rather, the computational methods used in
comparative genomics may take the lead by guiding traditional wet lab techniques such as gene22
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 23/29
knockout studies. Indeed, it is believed by many genomics researchers that one of the most im-
portant benefits of these computational methods is that they help molecular biologists in the
laboratory focus their research on those hypotheses that are most promising.
7. THE M ODULARITY OF THE GENOME
If we take common usage among researchers as definitive, then we would be forced to con-
clude immediately that genes exist. But the discussion from the previous sections has suggested
a more critical method for determining whether genes exist (and if so, what they are). That is,
we reinterpret the problem of the existence of genes as a problem of the referent of the term
‘gene’. With the problem formulated in such a way, two questions remain to be settled:
(1) Does the term ‘gene’ refer?
(2) If so, to what does the term ‘gene’ refer (to the best of our knowledge)?
We should note that these two questions are independent, in the sense that we may give a pos-
itive answer to the first without being able to answer the second. Also, it is important to note
that the first question belongs to the philosophy of language; in contrast, the second question
is a scientific one, which is philosophical only in that the philosophy of science should indicate
which empirical information bears upon it.
The previous discussion suggests that the best way to determine whether the term ‘gene’
refers is to look to the research programme within which the term is deployed. If there is an
ongoing research programme that is dedicated to discovering the characteristics of genes, and
that research is guided by the fact that particular causal powers are attributed to genes, then
we have good reason to hold onto the view that the term ‘gene’ refers. But if the research pro-
gramme has been abandoned, or if it has continued in name only – perhaps only by attributing
totally distinct causal powers to ‘genes’ – then we should hold (with Hull and Dupré) that genes
do not exist. For in such a case, the research programme has been abandoned, leaving behind
any available context upon which to fix the referent of the term.
So we ask what characteristics of genes are assumed by current research. When we consider
comparative genomics, the characteristic feature of this research that stands out is that it cru-
cially assumes that genes are, in an important sense, modular units on the chromosome. In
particular, we can identify the following features that genes are assumed to have, which weshall collectively label the ‘modularity of the genome’ hypothesis:
(1) Genes correspond to functional segments of nucleic acids on the chromosome.
(2) These sequences code for proteins, which perform identifiable functions – what we have
called ‘functional roles’.
(3) Genes tend to be conserved by natural selection – once a gene has evolved, it is likely to
be inherited by descendents of the originating species.23
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 24/29
(4) Genes are interchangeable modules – a gene may appear in one pathway of a particular
species, but be part of a different pathway in another species.
With the exposition of comparative genomics in section 6, it is easy to see that this research pro-
gramme crucially assumes the truth of theses (1) through (4). To see that it does in fact assume
the truth of these theses, we may briefly consider each in turn. Thesis (1) is obvious, since theannotation process assumes (as does everyone) that genes are to be identified by their location
on the chromosome. As for thesis (2), comparative genomics researchers must assume this as
a working hypothesis, or else it would be impossible to formulate a missing genes problem by
noting that a particular functional role has not been identified with a sequence of nucleic acids.
Genomicists assume the truth of thesis (3) in several ways; but most obviously, there would be
no reason to compare the annotations of several related species if there was no presumption
that these annotations would likely be shared by related species. And of course, the reason why
closely related species would be expected to have them in common is precisely because genes
(and their functional roles) are to be conserved by natural selection as species evolve. Lastly,
thesis (4) is assumed when comparative genomics researchers, in the course of investigating a
missing genes problem, look to related, but distinct, functional roles in other species.
Genes, then, are implicitly identified with a particular kind of sequence – namely, sequences
that are functional and modular in the sense given by theses (1) through (4), and whose modu-
larity is a product of evolution and natural selection.
At this relatively early stage of research into genomics, I am skeptical that it is possible to give
a more detailed characterization of the gene concept. But this should not be surprising – it is
only recently that large amounts of genomics data have become available, and this is a science
that is still in its infancy. And as I have noted above, it is perfectly ordinary that we may say
that a particular theoretical term refers, without being able to give it a full characterization. But
in spite of our inability to give a thorough intensional definition of the concept, there are im-
portant benefits to conceiving of genes as conserved, functional, modular sequences of nucleic
acids. In the remainder of this section, I shall briefly detail some of these benefits.
7.1. Evidence of genes. Given the complexity of the relationship between sequences andgenes,
one can hardly blame Duprè for announcing the end of the gene concept. However, as I have
argued, such pessimism is unwarranted. Indeed, it may be one of the more interesting corollar-
ies of the comparative genomics concept of the gene that it indicates what is right about these
earlier gene concepts. In particular, it shows us that these earlier gene concepts are evidence of
the existence of genes, although they cannot define what the gene concept is.
For example, consider (what has turned out to be) a naive hope that genes would correspond
to contiguous sequences of nucleic acids on the chromosome. Of course, we now recognize
that this is sometimes not the case. However, if we understand genes as evolved and conserved
functional modules on the chromosome, then it turns out that contiguity on the chromosome24
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 25/29
is (defeasible) evidence of the existence of genes. For the processes of recombination and other
genetic shuffling on the chromosome make it more likely that a sequence will be preserved
intact if it is not spread out over the chromosome. Thus, the fact that genes are functional mod-
ules implies that we would expect their physical characteristics to help them to be conserved
during those reshuffling processes. And indeed, as comparative genomics research has shown,it has turned out that genes often are contiguous for just this reason.
The lesson here is that we must not confuse evidential facts with definitional ones – in par-
ticular, the modularity of genes increases the probability that genes will be contiguous; thus,
the contiguity of an alleged gene is positive evidence that we have in fact identified a gene. But
like most evidential facts, these are defeasible. Some genes may be discontiguous, and yet be
functional modules. In general, when it turns out that a proposed mark of genes is found to not
hold generally, then we should not conclude that genes do not exist.
Indeed, the modular nature of genes shows why not only their contiguity, but also their loca-
tion on the chromosome, is evidential without being definitional. For the working hypothesisof comparative genomics is that genes are modular in at least two senses – for they not only are
functional modular units themselves, but they are embedded in a hierarchy of modules consist-
ing of functional roles, pathways, and subsystems. The fact that these ‘higher-level’ modules are
conserved by evolution and natural selection makes it the case that the location of genes occur-
ring in the same pathway are more likely to be located near each other for the same reason that
nucleic acids in the same gene are likely to be near each other. But again, this fact about the lo-
cation of genes on the chromosome does not serve as any part of the definition of what a gene
is; it is merely confirming evidence that particular sequences of nucleic acids are genes.
7.2. Why is it so difficult to characterize genes? It is an important virtue of this proposal that
it not only replaces some failed attempts to say what genes are, but that it also explains why it
is so difficult to characterize the gene concept in the first place. In fact, it is easy to see why the
gene concept is so elusive. For although modularity, as I have argued, is central to the nature of
genes, we do not yet understand the evolution of modularity.
Examples of evolved structures that display modularity are easy to come by. In philosophi-
cal literature, the best known discussion of modularity is undoubtedly the discussion that was
instigated by Jerry Fodor regarding the modularity of mind [4]. Other examples are less well-
known in philosophical discussions. For instance, recent research in the nascent field of neu-
roeconomics has uncovered neural structures that appear to function as discrete modules (e.g.
see [1, 6, 7]). And it has been well-known in computer science that when neural networks are
subject to evolutionary pressures through so-called ‘genetic algorithms’, it is common for the
resulting structures to exhibit modularity (e.g. [12–14]).
If the research methodology of comparative genomics is borne out in the long run (as I be-
lieve it will be) then it will turn out that modularity has evolved not only in gross anatomical25
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 26/29
structures, but in the genetic code as well. Thus, to see why it is so difficult to characterize the
nature of genes, we should see how this problem is an instance of a more general problem that
is extremely difficult. Let us call this ‘the problem of evolved modularity’.
The problem of evolved modularity has been addressed in the philosophical literature, but
not in a technically satisfying way. For example, Günther Wagner has discussed two major pro-cesses that may bring about the evolution of modularity, which he calls ‘parcellation’ and ‘inte-
gration’ [30, p. 38]. Applying these concepts to genes, ‘parcellation’ refers to the ‘elimination of
pleiotropic effects’ between different sets of genes or nucleic acid sequences and the ‘mainte-
nance and/or augmentation of pleitropic effects’ within genes or nucleic acid sequences. The
concept of integration is concerned with the construction of higher-level modularity; it is the
‘creation of pleiotropic effects’ among genes. Thus, in the context of the evolution of genes
and pathways, if we consider genes to be the lowest level in a hierarchy of modularity, parcel-
lation would be a general term referring the processes whereby the modularity of the gene is
produced. At a higher level of modularity, integration is the general process whereby genesbecome organized into pathways.
It should not be controversial at all that processes of parcellation and integration must take
place in the evolution of genes, pathways, and higher levels of modularity. Indeed, these terms,
as defined by Wagner, are so general that almost no substantive empirical claim is made by
asserting that these processes take place. The interesting challenge, which may be framed in
terms of these two processes, is therefore to determine by what evolutionary mechanisms par-
cellation and integration do take place. And it is here that comparative genomics is extremely
useful. For as I have outlined in above, there is a useful positive feedback loop between genome
annotation and the discovery of phylogenetic history. Genome annotation – as it is practiced incomparative genomics – depends crucially on our having at least a partial phylogenetic history
of the species, because the technique requires comparisons among more or less closely related
species. Conversely, when a set of annotations is completed, reference to existing sequence
data for other species may suggest phylogenetic relationships that have been unknown. Thus,
as more sequence data and annotated sequences become available, we are able to look farther
back in the evolutionary history of the species. In fact, this process may allow us to reconstruct
how the gene arose in the first place, and learn about the timing and process whereby genes
became organized into particular pathways. It is important to note that this is not merely spec-
ulation; in an increasing number of cases, this has been accomplished.10
10For example, comparative genomics has made is possible to reconstruct the evolutionary origin of the Prosthe-
cobacter tubulin genes [11], lysine biosynthesis [17], as well as specific functional roles in pathways in the lysine
biosynthesis subsystem [29]. An informative discussion of methodology may be found in [31].
26
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 27/29
This positive feedback loop between phylogenetic inference and genome annotation also
makes contact with the distinctively philosophical problem of analyzing the gene concept. Be-
cause it is essential to the gene that it is an evolved modular structure, a fully satisfactory ac-
count of the gene will require an understanding of how such modular structures evolve. At this
time, we can only gesture at the mechanisms by which modularity evolves; but comparativegenomics will allow us to learn how modularity arises (when it does). At that point, we will be
able to offer a specific, etiological account of the gene.
8. CONCLUSION
Although it is a significant amount of work to get clear on the research methodology of com-
parative genomics, there is more than enough philosophical payoff for doing so. In particular,
it turns out that the fact about genes that is crucial to understanding comparative genomics
is that this research must assume that genes are modular. Genes are conceived of as discrete,
functional units that are interchangeable among various pathways and subsystems, and whichare also conserved by evolution. If my arguments are correct, then it turns out that the various
alleged features of genes (such as contiguity, location on the chromosome, etc.) that have been
seized upon as providing essential features of genes are actually by-products of the modularity
of genes.
If the arguments in this paper have been correct, then the most significant payoff of the cur-
rent study might not be a positive characterization of the gene, but instead the identification of
a worthwhile and neglected research problem. For we will not be able to provide a fully ade-
quate gene concept without first understanding the evolution of modularity. If we were to have
an adequate theory of the evolution of modularity, other philosophical problems would be elu-cidated; these include the modularity of the mind and perhaps the units of selection problem.
Fortunately, comparative genomics is beginning to provide valuable empirical data on how a
complex modular structure has evolved. Thus, the problem of characterizing the gene con-
cept may be a route to understanding other philosophical problems that may be illuminated
through a better understanding of modularity.
27
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 28/29
REFERENCES
1. Colin Camerer, George Loewenstein, and Drazen Prelec, Neuroeconomics: How neuroscience can inform eco-
nomics , Journal of Economic Literature 43 (2005), 9–64.
2. Berent Enç, Reference of theoretical terms , Noûs 10 (1976), no. 3, 261–282.
3. Zachary Ernst, Philosophical issues arising from genomics , Oxford Handbook of Philosophy of Biology (Michael
Ruse, ed.), Oxford University Press, 2008.
4. J.A. Fodor, The modularity of mind , MIT Press Cambridge, MA, 1983.
5. Alan Garfinkel, Reductionism , The Philosophy of Science (Richard Boyd, Philip Gasper, and J.D. Trout, eds.),
MIT Press, 1991, pp. 443–459.
6. Paul W. Glimcher, Decisions, uncertainty, and the brain: The science of neuroeconomics , MIT Press, Cambridge,
Massachusetts, 2003.
7. Paul W. Glimcher and Aldo Rustichini, Neuroeconomics: The consilience of brain and decision , Science 306
(2004), 447–452.
8. David L. Hull, Informal aspects of theory reduction , Philosophy of Science Association (1974), 653–670.
9. D.L. Hull, Reduction in Genetics–Biology or Philosophy? , Philosophy of Science 39 (1972), no. 4, 491–499.
10. N. Ivanova, A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik,
N. Mikhailova, A. Lapidus, et al., Genome sequence of Bacillus cereus and comparative analysis with Bacillus
anthracis , Nature 423 (2003), no. 6935, 87–91.
11. Cheryl Jenkins, Ram Samudrala, et al., Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthe-
cobacter , Proceedings of the National Academy of Sciences of the United States of America 99 (2002), 17049–
17054.
12. Nadav Kashtan and Uri Alon, Spontaneous Evolution of Modularity and Network Motifs , Proceedings of the
National Academy of Sciences of the United States of America 102 (2005), no. 39, 13773–13778.
13. B. Kosko, Hidden patterns in combined and adaptive knowledge networks , International Journal of Approxi-
mate Reasoning 2 (1988), no. 4, 377–393.
14. , Neural networks and fuzzy systems: a dynamical systems approach to machine intelligence , Prentice-Hall, 1992.
15. Saul Kripke, Naming and necessity , Harvard University Press, Cambridge, 1980.
16. Frederick W. Kroon, Theoretical terms and the causal view of reference , Australasian Journal of Philosophy 63
(1985), no. 2, 143–166.
17. Hiromi Nishida, Makoto Nishiyama, Nobuyuki, Takehide Dosuge, Takayuki Hoshino, and Hisakazu Yamane, A
Key to the Evolution of Amino Acid Biosynthesis , Genome Research 9 (1999), 1175–1183.
18. Robert Nola, Fixing the reference of theoretical terms , Philosophy of Science 47 (1980), no. 4, 505–531.
19. R. Overbeek, M. Fonstein, M. D’Souza, G.D. Pusch, and N. Maltsev, The use of gene clusters to infer functional
coupling , Proc Natl Acad Sci US A 96 (1999), no. 6, 2896–2901.
20. Ross Overbeek, Genomics: what is realistically achievable? , Genome Biology 1 (2000), 1–3.
21. Ross Overbeek, Terry Disz, and Rick Stevens, The SEED: A peer-to-peer environment for genome annotation ,
Communications of the Association for Computing Machinery 47 (2004), 46–51.
22. Ross Overbeek et al., The ERGO genome analysis and discovery system , Nucleic Acids Research 31 (2003), no. 1,
164–171.
23. , The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes ,
Nucleic Acids Research 33 (2005), no. 17, 5691–5702.
24. Hilary Putnam, Meaning and Reference , The Journal of Philosophy 70 (1973), no. 9, 699–711.
28
8/12/2019 Ernst - Unknown - Comparative Genomics and the Gene Concept
http://slidepdf.com/reader/full/ernst-unknown-comparative-genomics-and-the-gene-concept 29/29
25. Willard Van Orman Quine, Reference and modality , From a Logical Point of View, Harvard University Press,
1953.
26. Alexander Rosenberg, Instrumental biology or the disunity of science , University of Chicago Press, Chicago,
1994.
27. Elliott Sober, Reconstructing the past: Parsimony, evolution, and inference , MIT Press, Cambridge, Mas-
sachusetts, 1988.
28. P. Kyle Stanford and Philip Kitcher, Refining the causal theory of reference for natural kind terms , Philosophical
Studies 97 (2000), 99–129.
29. A.M. Velasco, J.I. Leguina, and A. Lazcano, Molecular Evolution of the Lysine Biosynthetic Pathways , Journal of
Molecular Evolution 55 (2002), 445–459.
30. Günther Wagner, Homologues, Natural Kinds and the Evolution of Modularity , American Zoologist 36 (1996),
36–43.
31. Itai Yanai and Charles DeLisi, The society of genes: networks of functional links between genes from comparative
genomics , Genome Biology 3 (2002), no. 11, 1–12.
E-mail address : [email protected]
DEPARTMENT OF PHILOSOPHY , UNIVERSITY OF M ISSOURI-C OLUMBIA
URL : www.missouri.edu/~ernstz