271===Chromosome Numbers S.lepidophyla===
-
Upload
sellaginella -
Category
Documents
-
view
220 -
download
0
Transcript of 271===Chromosome Numbers S.lepidophyla===
-
8/7/2019 271===Chromosome Numbers S.lepidophyla===
1/6
Variation in Chromosome Numbers, CMA Bands and 45S rDNA Sites in
Species ofSelaginella (Pteridophyta)
A D R I A N A B U A R Q U E M A R C O N , I V A C A R N E I R O L E A O B A R R O S a nd MA R C E L O G U E R R A *
Universidade Federal de Pernambuco, Centro de Ciencias Biologicas, Departamento de Botanica,
Rua Nelson Chaves s/n, Cidade Universitaria, 50.670-420, Recife, Pernambuco, Brasil
Received: 16 January 2004 Returned for revision: 1 May 2004 Accepted: 15 August 2004 Published electronically: 26 November 2004
Background and Aims Selaginella is the largest genus of heterosporous pteridophytes, but karyologically the genusis known only by the occurrence of a dysploid series of n = 712, and a low frequency of polyploids. Aiming tocontribute to a better understanding of the structural chromosomal variability of this genus, different stainingmethods were applied in species with different chromosome numbers. Methods The chromosome complements of seven species of Selaginella were analysed and, in four of them, thedistribution of 45S rDNA sites was determined by fluorescent in situ hybridization. Additionally, CMA/DA/DAPIand silver nitrate staining were performed to investigate the correlation between the 45S rDNA sites, the hetero-
chromatic bands and the number of active rDNA sites. Key Results The chromosome numbers observed were 2n = 18, 20 and 24. The species with 2n = 20 exhibitedchromosome complement sizes smaller and less variable than those with 2n = 18. The only species with 2n = 24,S. convoluta, had relatively large and asymmetrical chromosomes. The interphase nuclei in all species were of thechromocentric type. CMA/DA/DAPI staining showed only a weak chromosomal differentiation of heterochromaticbands. In S. willdenowii and S. convoluta eight and six CMA+ bands were observed, respectively, but no DAPI+
bands. The CMA+ bands corresponded in number, size and location to the rDNA sites. In general, the number ofrDNA sites correlated with the maximum number of nucleoli per nucleus. Ten rDNA sites were found in S. plana(2n = 20), eight in S. willdenowii (2n = 18), six in S. convoluta (2n = 24) and two in S. producta (2n = 20). Conclusions The remarkable variation in chromosome size and number and rDNA sites shows that dramatickaryological changes have occurred during the evolution of the genus at the diploid level. These data further suggestthat the two putative basic numbers of the genus, x = 9 and x = 10, may have arisen two or more timesindependently. 2004 Annals of Botany Company
Key words: Pteridophyta, Selaginella, chromosome number, fluorochrome staining, silver nitrate staining, in situhybridization, 45S rDNA.
I N T R O D U C T I O N
Selaginella P. Beauv. is a heterosporous genus of the familySelaginellaceae, comprising approx. 750 species distributedthroughout the tropical regions, including 250 species in theAmericas (Tryon and Tryon, 1982; Kramer and Green,1990). They are generally found in humid and shady forests,although they can also be present in dry forests, swamps, ondamp rocks along riverbanks or near waterfalls, and even incold regions such as the Alps, or in rocky deserts (Tryon andTryon, 1982).
Polyploidy and hybridization have occurred numerous
times in the evolution of the pteridophytes, resulting inthe characteristically high chromosome numbers seen inthis group (Walker, 1984). Nonetheless, high chromosomenumbers are rare among the heterosporous pteridophytes.Selaginella, the largest heterosporous genus, has only a fewpolyploid species, although a large dysploid variation hasbeen reported. The chromosome number ranges between2n = 14 and 2n = 60, and different authors have reporteddifferent basic chromosome numbers for the genus: x = 7,8, 9, 10, 11 and 12 (Kuriachan, 1963; Jermy et al., 1967;Takamiya, 1993).
In spite of several cytotaxonomical studies, the karyolo-gical evolution of this group remains poorly understood.Cytological investigations have been based primarily onchromosome counts and, in a few cases, on chromosomemorphology. Meanwhile, in angiosperms and gymnospermsthe karyological studies in the last two decades have turnedto more specific methods such as banding methods andfluorescent in situ hybridization (FISH) (Greilhuber,1995; Stace and Bailey, 1999). Heterochromatic bands inplants have been analysed mainly by C-banding or bybase-specific fluorochrome staining (Guerra, 2000).Fluorochrome banding has the advantage of being a simpler,
more reproducible and less destructive method, as com-pared with C-banding (Guerra, 1993). The fluorochromeschromomycin A (CMA) and 40,6-diamidino-2-phenylindole(DAPI) exhibit preferential staining for GC and AT-richDNA sequences, respectively, allowing the identificationof different types of heterochromatin. Distamycin A(DA) has also been used to enhance the contrast betweenCMA and DAPI (Schweizer and Ambros, 1994; Marconet al., 2003). The 45S rDNA sites are generally positivelystained with CMA and negatively stained with DAPI. Inmany species, the rDNA sites are the only positively stainedCMA bands (see, for example, Melo and Guerra, 2003). Thevariation in number and position of the rDNA sites has been
Annals of Botany 95/2 Annals of Botany Company 2004; all rights reserved
* For correspondence. E-mail [email protected]
Annals of Botany 95: 271276, 2005
doi:10.1093/aob/mci022, available online at www.aob.oupjournals.org
-
8/7/2019 271===Chromosome Numbers S.lepidophyla===
2/6
demonstrated to be an additional important karyotypefeature to the cytotaxonomic analyses of some angiospermgenera, such as Clivia (Ran et al., 1999) and Sanguisorba(Mishima et al., 2002). Silver nitrate staining is anothermethod that allows the visualization of nucleolus organizerregions (NORs), which are also related to chromosomalsecondary constrictions and rDNA sites, but after the silvernitrate staining only those sites activated in an interphasenucleus are stained in the subsequent prophase or metaphase(Moscone et al., 1995). In general, the maximum number ofsilver-stained sites in prophase or metaphase chromosomesand the maximum number of nucleoli in interphase nucleicorrespond to the number of 45S rDNA sites (see, for exam-
ple, Moscone et al., 1995). This relationship seems to betrue also for homosporous pteridophytes (Kawakami et al.,1999; Marcon et al., 2003).
In order to better understand the cytogenetics and evolu-tion of this genus, the chromosome numbers were analysedin seven Brazilian species ofSelaginella and in four of themthe distribution of 45S rDNA sites was observed. Addition-ally, the heterochromatic blocks stained with CMA/DA/DAPI and the number of nucleoli per nucleus were inves-tigated, as an indication of the number of NORs. The resultsare discussed in relation to the chromosomal evolution ofthe genus.
M A T E R I A L S A N D M E T H O D S
The species investigated, with their respective collectionsites, chromosome numbers and previous chromosomecounts are listed in Table 1. Part of the material collectedwas prepared for the herbarium, and the vouchers weredeposited in the UFP herbarium (Federal University ofPernambuco, Brazil). Another part of the material wasmaintained at the experimental garden of the Departmentof Botany, at the Federal University of Pernambuco, forcytogenetic analysis.
Leaf buds were collected and pretreated in 0002 M8-hydroxyquinoline for 1 h at room temperature, followed
by 23 h at 10 C. Subsequently, they were fixed in Carnoy(ethanol : acetic, acid 3 : 1) for 24 h at room temperature, andstored at 20 C.
For conventional chromosome staining, the fixed leafbuds were washed twice in distilled water for 5 min, hydro-lysed in 5 N HCl for 30 min, and then the meristem wasisolated and squashed in 45 % acetic acid (Guerra, 1983).The preparation was stained with Giemsa 5 % and mountedwith Entellan. Chromosome measurements were performedon amplified photographs with the aid of a pachymeter.
The triple staining with the fluorochromes CMA, DAPIand DA was performed according to Schweizer and Ambros(1994). Leaf buds were washed twice in distilled water for
5 min, digested in 2 % cellulase/20 % pectinase for 2 h at37 C, and squashed in 45 % acetic acid. After coverslipremoval the slides were aged for 3 d at room temperature,stained with CMA (05 mg mL1, 1 h), counterstained withdistamycin A (01 mg mL1, 30 min), stained with DAPI(2 mg mL1, 30 min) and mounted in McIlvaines (pH 70)bufferglycerol (v/v, 1 : 1) containing 25 mM MgCl2.CMA/DAPI staining without distamycin A produced apoorer differentiation.
Leaf buds without pretreatment were fixed directly intoCarnoy and used for silver nitrate staining. Slides wereprepared using the same enzyme treatment employed forfluorochromes, except that they were not aged before stain-ing. A small drop of 50 % silver nitrate diluted in formic
acid was placed over the cells, covered with a coverslip andincubated in a moist chamber at 60 C (Rufas et al., 1987)for approx. 10 min or until adequate staining was attained.
To locate the 45S rDNA sites, probes SK18S and SK25Swere used, containing 18S and 25S rDNA of Arabidopsisthaliana (Unfried etal., 1989; Unfried and Gruendler,1990),kindly provided by Prof. D. Schweizer of the Universityof Vienna, and labelled by nick translation with biotin-11-dUTP (Sigma) or digoxigenin-11-dUTP (Roche). Theprobe was detected with mouse anti-biotin monoclonalantibody (Dakopatts no. M743) and visualized with rabbitanti-mouse antibody conjugated to tetramethyl rhodamineisothiocyanate (TRITC) (Dakopatts no. R270) or detected
T A B L E 1. Species ofSelaginella analysed, with their respective voucher number, provenance, chromosome number and previouscounting
SpeciesVouchernumber Provenance
Chromosomenumber (2n)
Previouscountings (2n) References
S. muscosa Spring 36588 Bonito 18 S. simplex Baker 36412 Gravata 18 S. willdenowii
(Desv. ex Poiret) Baker36589 Recife 18 18 Jermy et al. (1967); Fabri (1963);
Kuriachan (1963)Selaginella sp. Itamaraca 18 S. producta Baker 36410
36411PaulistaIgarassuu
2020
36591 Bonito 20 S. plana (Desv. ex Poiret)Hieron
36409 Recife 20 20 Jermy et al. (1967); Fabri (1963);Kuriachan (1963)
36590 Rio de Janeiro 20 20 Jermy et al. (1967); Fabri (1963);Kuriachan (1963)
S. convoluta (Arnott) Spring 36408 Petrolina 24
All the samples were collected in the Brazilian state of Pernambuco, except S. plana, which is from the state of Rio de Janeiro.
272 Marcon et al. Cytogenetic Variability in Species of Selaginella
-
8/7/2019 271===Chromosome Numbers S.lepidophyla===
3/6
with sheep anti-digoxigenin antibody conjugated tofluorescein isothiocyanate (FITC) (Boehringer Mannheimno.1207741) and FITC-conjugated rabbit anti-sheep(Dakopatts F135, DAKO). The technique was based onMoscone etal.(1996),withsomemodifications(denaturationat 80 C and the post-hybridization washes in 01 SSC at42 C). The hybridization mixture contained 60 % forma-mide, 5 % dextran sulfate, 2 SSC, 001 % salmon spermDNA,andprobeatafinalconcentrationof1230 ngmL1. Inmost hybridization experiments, a 5S rDNA probe obtainedfrom total genomic DNAofPassiflora edulisSims (Melo and
Guerra,2003)wasaddedtothemixturebutnoclearsignalwasobserved. Slides were stained with DAPI (2 mg mL1) andmounted in Vectashield (Vector).
The best cells were captured with a CCD Cohu camera ona Leica DMLB microscope, or photographed using KodakImagelink HQ ASA 25 film for bright field, or Kodak ASA400 film for fluorescence photography.
R E S U L T S
The seven species of Selaginella analysed in the presentwork showed the following chromosome numbers: 2n = 18
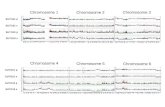
[S. muscosa Spring, S. simplex Baker, S. willdenowii(Desv. ex Poiret) Baker and Selaginella sp.], 2n = 20[S. producta Baker and S. plana (Desv. ex Poiret) Hieron.]and 2n = 24 [(S. convoluta (Arnott) Spring] (Fig. 1AG).
Among the species with 2n = 18, S. willdenowii andSelaginella sp. exhibited the largest chromosomes, mostof them meta- to submetacentric. The chromosomes inS. willdenowii varied between 144 and 248 mm (Fig. 1A),and showed up to six satellites: two on the short arms andfour on the long arms of submetacentric chromosomes. InSelaginella sp.the chromosome size variedbetween 135and
232 mm and three satellites were observed: two on a meta-centric pair, and a third on a submetacentric chromosome(Fig. 1B). On the other hand, S. simplex and S. muscosa hadmuch smaller chromosomes, varying, respectively, between085 and 126 mm and 080 and 162 mm (Fig. 1C and D).
The species with 2n = 20 had smaller chromosomes,ranging from 070 to 104 mm in S. plana, and 078 to143 mm in S. producta (Fig. 1E and F). The position ofthe centromere could not be observed in any of these fourspecies, mainly due to the small size of their chromosomes.Satellites were also not clearly identified.
In the only species with 2n = 24, S. convoluta, themorphology of the chromosomes seemed to vary from
A
E
J K L
F G H I
B C D
F I G . 1. Chromosome complement (AG), interphase nuclei (H and I) and number of NORs and nucleoli (JL) observed in species of Selaginella:(A) S. willdenowii (2n = 18) with two satellites (arrows); (B) Selaginella sp. (2n = 18) with three satellites (arrows); (C) S. simplex (2n = 18);(D) S. muscosa (2n = 18); (E) S. plana (2n = 20); (F) S. producta (2n = 20); (G) S. convoluta (2n = 24); (H and I) chromocentric interphase nuclei of
S.plana and S. willdenowii, respectively; (J) prometaphasecell showingNORs (arrows); (K and L) prometaphasecells showingchromosomes associated withnucleoli (arrows) in S. plana (K) and S. convoluta (L). The bar in A = 5 mm.
Marcon et al. Cytogenetic Variability in Species of Selaginella 273
-
8/7/2019 271===Chromosome Numbers S.lepidophyla===
4/6
metacentric to acrocentric. Chromosome size variedbetween 070 and 177 mm and satellites were not observed(Fig. 1G).
In all species analysed, the interphase nucleus structurewas granulated, with small chromocentres, corresponding tothe chromocentre type, according to the classificationof Tanaka (1971). Some species showed larger and moresharply outlined chromocentres (Fig. 1H and I).
Staining with silver nitrate revealed that the maximumnumber of nucleoli per nucleus varied among species(Table 2). In S. producta the maximum number of nucleoliper nucleus was two, whereas in the other three speciesinvestigated, the maximum number ranged between sixand ten, although most nuclei exhibited only three nucleoli.Selaginella willdenowii exhibited a variation of one to tennucleoli per interphase nucleus and up to ten NORs wereobserved in prometaphase (Fig. 1J). In S. convoluta andS. plana, it was not possible to identify NORs, but six toten prophase or prometaphase chromosomes associatedwith nucleoli were observed (Fig. 1K and L).
A high number of metaphase cells was obtained from
S. willdenowii, S. plana, S. producta and S. convoluta mak-ing it possible to analyse the chromosomes with fluoro-chromes and in situ hybridization. Staining with CMA/DA/DAPI revealed differentiation of bands only inS. willdenowii and S. convoluta. The chromosomes of theother species stained deeply and homogeneously with bothCMA and DAPI, even when counterstained with distamy-cin A. In prometaphase cells of S. willdenowii, up to eightCMA+/DAPI terminal bands were observed (Fig. 2A andB). In S. convoluta (2n = 24), six terminal CMA+/DAPI
bands were localized in three chromosome pairs (Fig. 2Dand E). No DAPI+ bands were observed in these two spe-cies, while the CMA+ bands were often negatively stainedwith DAPI.
In situ hybridization of 45S rDNA in chromosomes ofS. willdenowii revealed the presence of terminal sites on thelong arms of two chromosome pairs and on the short arms ofanother two chromosome pairs (Fig. 2C). In two populationsofS. plana analysed, five submetacentric pairs were labelledterminally: three pairs on the long arms and two pairs on theshort arms (Fig. 2H and I). While in one of the populations itwas not possible to observe clearly the smaller site, for thisstains weakly, in the another population analysed the tensites of 45S rDNA were clearly labelled (Fig. 2I). InS. convoluta there were four sites on two apparently acro-centric pairs and two sites on a submetacentric pair (Fig.2F).In S. producta (Fig. 2G and J) only one chromosome pair was
labelled, at the terminal position, in the two populations.These two signals were always remarkably distinct in inter-phase nuclei (Fig. 2K), whereas up to ten signals wereobserved in the interphase nuclei of S. plana (Fig. 2L).
D I S C U S S I O N
The chromosome numbers and the structure of interphasenuclei reported in the present work are compatible withthe karyological data known for the genus Selaginella(Kuriachan, 1963; Jermy et al., 1967; Ghatak, 1977;Takamiya, 1993). Of the seven species examined, onlyS. plana and S. willdenowii had been analysed previously.The chromosome numbers observed for these speciesagreed with the previous counts.
With regard to chromosome size and morphology, thereis a strong trend to conserve the karyotype symmetry, withpredominance of metacentric and submetacentric chromo-somes. The species with 2n = 20 analysed in this samplewere more stable in chromosome size than those with 2n =18. Takamiya (1993) found similar results among species
with x = 10 and x = 9. A higher number of meta- andsubmetacentric chromosomes was observed in the karyo-type of all species analysed, and appears to be a commontrend in this genus (Takamiya, 1993), and in other hetero-sporous pteridophytes (Kuriachan, 1979, 1994). On theother hand, acro- and telocentric chromosomes are morefrequent among homosporous pteridophytes (Kawakami,1982; Takamiya et al., 1992; Marcon et al., 2003).
In situ hybridization with 45S rDNA revealed the mostprominent structural variation between species. One specieswith 2n = 20, S. plana, displayed ten sites of 45S rDNAwhereas another one with the same chromosome number,S. producta, exhibited only two. On the other hand,S. willdenowii with 2n = 18, showed more rDNA sites
(eight) than S. convoluta (six) with 2n = 24. In angiospermsa similar variation has been widely reported. In Passiflora,for instance, the number of 45S rDNA sites varied from oneto three pairs among diploid species (Melo and Guerra,2003). In Paeonia (Zhang and Sang, 1999), five diploidspecies (2n = 10) exhibited three to ten pairs of rDNAsites. Variation in the number of rDNA sites among angios-perm species of the same ploidy level has been attributed tochromosome rearrangements, transpositional events andgene silencing (Moscone et al., 1999). Likewise, similarmechanisms may be acting in Selaginella species.
In S. plana, S. convoluta and S. producta there wasa perfect correlation between the maximum number
T A B L E 2. Variation in number of chromosomes, rDNA sites and nucleoli per nucleus in four species of Selaginella
Variation in the number of nucleoli per nucleus
Species 2n Number of rDNA sites No. of cells analysed Range Most frequent numbers
S. willdenowii 18 8 1015 110 3 (29.5 %) and 4 (21.1 %)
S. plana 20 10 800 110 2 (27.8 %) and 3 (31.7 %)S. producta 20 2 308 12 1 (94.2 %)S. convoluta 24 6 528 16 2 (28.0 %) and 3 (35.7 %)
274 Marcon et al. Cytogenetic Variability in Species of Selaginella
-
8/7/2019 271===Chromosome Numbers S.lepidophyla===
5/6
of nucleoli (10, 6 and 2, respectively) and the number of 45SrDNA sites. The wide variation in the number of nucleoliper nucleus observed in all species examined, with a higherfrequency of nuclei with few nucleoli, is probably due to the
fusion of nucleoli, as observed in angiosperms (Mosconeet al., 1995).
Curiously, S. willdenowii, although having up toten NORs, displayed only eight 45S rDNA sites and six
A
D
G
J K L
H I
E F
B C
F I G . 2. Distribution of theCMA/DAPIbands and45S rDNAsitesin speciesofSelaginella: S. willdenowii (Aand B)and S. convoluta (D andE), stained withCMA (A and D) and DAPI (B and E),respectively. Arrows in A,B, D and E indicate the CMA+/DAPI bands. 45S rDNA sites (C and FL) in S. willdenowii
(C), S. convoluta (F), in twopopulationsofS. producta (G andJ), andin twopopulationsofS.plana (Hand I).Interphase nucleiofS. producta (K),showingthetwo sites of 45S rDNA and of S. plana (L), showing up to ten sites. The bar in L = 5 mm.
Marcon et al. Cytogenetic Variability in Species of Selaginella 275
-
8/7/2019 271===Chromosome Numbers S.lepidophyla===
6/6
chromosomes with satellites. This apparent lack of correla-tion between NORs and rDNA sites may be due to a pair ofrDNA sites very reduced in size, not detected by FISH. Thenumber of satellites observed, on the other hand, is oftenlower than the number of rDNA sites (see, for example,Guerra et al., 1996), and in some cases they are not observed
at all, as in S. convoluta, S. plana and S. producta.The CMA/DA/DAPI staining revealed only CMA+/DAPI
bands in S. willdenowii and S. convoluta. The number ofCMA+ bands was positively correlated with the number ofrDNA sites and the maximum number of nucleoli observedin these species. Similar results were found in the homo-sporous genus Acrostichum (Marcon et al., 2003), suggest-ing that these three features are also positively correlated inpteridophyte chromosomes. No previous report of CMAbands in pteridophytes was found in the literature.
Jermy et al. (1967) proposed that the primary basic num-ber ofSelaginella would be x = 10, characteristic of speciesgrowing in dense tropical and subtropical forests (see alsoKuriachan, 1963). However, later karyological analyses(Takamiya, 1993), as well as morphological, anatomicaland paleobotanical data (Mukhopadhyay, 1998) suggestthat the basic number of the genus would be x = 9, gen-erating successive dysploids with n = 10, 11 and 12 on onehand and n = 8 and 7 on the other. Takamiya (1993) pro-posed that dysploidy might have occurred repeatedly indifferent evolutionary lines within each subgenus. Indeed,the remarkable variation in chromosome number and in sizeand number of rDNA sites, seems to indicate that structuralvariation is rather common in the genus and that itskaryological evolution may be far more complex, witheach basic number arising two or more times.
A C K N O W L E D G E M E N T S
The authors are very greatful to Dr Ivan Valdespino fromUniversity of Panama, for species identification, and toConselho Nacional de Desenvolvimento Cientfico eTecnologico (CNPq) and Fundacao de Amparo a Cienciae Tecnologia do Estado de Pernambuco (FACEPE) forfinancial support.
L I T E R A T U R E C I T E D
Fabri F. 1963. Primo supplemento alle tavole cromosomiche dellePteridophyta di Alberto Chiarugi. Cariologia 16: 237335.
Ghatak J. 1977. Biosystematic survey of pteridophytes from ShevaroyHills, South India. Nucleus 20: 105108.
Greilhuber J. 1995. Chromosomes of the monocotyledons (generalaspects). In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ, eds.
Monocotyledons: systematics and evolution. Kew: Royal BotanicGardens, 379414.
Guerra M. 1983. O uso de Giemsa na citogenetica vegetalcomparacaoentre a coloracao simples e o bandeamento. Ciencia e Cultura35: 191193.
Guerra M. 1993. Cytogenetics of Rutaceae. V. High chromosomalvariability in Citrus species revealed by CMA/DAPI staining.
Heredity 71: 234241.Guerra M. 2000. Patterns of heterochromatin distribution in plant chromo-
somes. Genetics and Molecular Biology 23: 10291041.Guerra M, Kenton A, Bennett MD. 1996. rDNA sites in mitotic and
polytene chromosomes of Vigna unguiculata (L.) Walp. andPhaseolus coccineus L. revealed by in situ hybridization. Annals of
Botany 78: 157161.
Jermy AC, Jones K, Colden C. 1967. Cytomorphological variation inSelaginella. Botanical Journal of the Linnean Society 60: 147158.
Kawakami S. 1982. Karyomorphological studies on Japanese Pteridaceae.IV. Discussion. Bulletin of Aichi University of Education (Natural
Science) 31: 175186.Kawakami SM, Kondo K, Kawakami S. 1999. Analysis of nucleolar
organizer constitution by fluorescent in situ hybridization (FISH)
in diploid and artificially produced haploid sporophytes of the fernOsmunda japonica (Osmundaceae). Plant Systematics and Evolution216: 325331.
Kramer KU, Green PS. 1990. The families and genera of vascular plants.Vol. I. Pteridophytes and gymnosperms. Berlin: Springer-Verlag.
Kuriachan PI. 1963. Cytology of the genus Selaginella. Cytologia 28:376380.
Kuriachan PI. 1979. Interspecific origin of Salvinia molesta Mitchell:evidence from karyotype. Indian Journal of Botany 2: 5154.
Kuriachan PI. 1994. Karyotype of Regnellidium diphyllum Lindm.Caryologia 47: 311314.
Marcon AB, Barros ICL, Guerra M. 2003. A karyotype comparisonbetween two closely related Acrostichum L. (Pteridaceae) species.
American Fern Journal 93: 116125.Melo NF, Guerra M. 2003. Variability of the 5S and 45S rDNA sites
in Passiflora L. species with distinct base chromosome numbers.Annals of Botany 92: 309316.
Mishima M,Ohmido N, Fukui K, Yahara T. 2002. Trends in site numberchange of rDNA loci during polyploid evolution in Sanguisorba(Rosaceae). Chromosoma 110: 550558.
Moscone EA, Klein F, Lambrou M, Fuchs J, Schweizer D. 1999.Quantitative karyotyping and dual-color FISH mapping of 5S and18S-25S rDNA probes in the cultivated Phaseolus species(Leguminosae). Genome 42: 12241233.
Moscone EA, Loidl J, Ehrendorfer F, Hunziker AT. 1995. Analysisof active nucleolus organizing regions in Capsicum (Solanaceae) bysilver staining. American Journal of Botany 82: 276287.
Moscone EA, Matzke MA, Matzke AJM. 1996. The use of combinedFISH/GISH in conjunction with DAPI counterstaining to identifychromosomes containing transgene inserts in amphidiploid tobacco.Chromosoma 105: 231236.
Mukhopadhyay R. 1998. Cytotaxonomic observations on SelaginellaBeauv. Phytomorphology 48: 343347.
Ran Y, Murray BG, Hammett KRW. 1999. Karyotype analysis ofthe genus Clivia by Giemsa and fluorochrome banding and in situhybridization. Euphytica 106: 139147.
Rufas JS, Gimenez-Abian J, Suja JA, Garcia De La Vega C. 1987.Chromosome organization in meiosis revealed by light microscopeanalysis of silver-staining cores. Genome 29: 706712.
SchweizerD, Ambros PF. 1994. Chromosome banding. In: Gosden JR, ed.Methods in molecular biology, Vol.29. Totowa: Humana Press, 97112.
Stace CA, Bailey JP. 1999. The value of genomic in situ hybridization(GISH)in planttaxonomicand evolutionarystudies. In: HollingsworthPM, Bateman RM, Gornall RJ, eds. Molecular systematics and plantevolution. London: Taylor & Francis, 199210.
Takamiya M. 1993. Comparative karyomorphology and interrelationshipsof Selaginella in Japan. Journal of Plant Research 106: 149166.
Takamiya M, Osato K, Ono K. 1992. Karyomorphological studies onWoodwardia sensu lato of Japan. Botanical Magazine 105: 247263.
Tanaka R. 1971. Types of resting nuclei in Orchidaceae. Botanical
Magazine 84: 118122.Tryon RM, Tryon AF. 1982. Ferns and allied plantswith special
reference to tropical America. New York: Springer-Verlag.Unfried I, Gruendler P. 1990. Nucleotide sequence of the 58S and 25S
rRNA genes and of the internal transcribed spacers from Arabidopsis
thaliana. Nucleic Acids Research 18: 4011.Unfried I, Stocker U, Gruendler P. 1989. Nucleotide sequence of the 18S
rRNA gene from Arabidopsis thaliana Co10. Nucleic Acids Research
17: 7513.Walker TG. 1984. Chromosomes and evolution in pteridophytes. In:
Sharma AK, Sharma A, eds. Chromosome evolution of eukaryoticgroups. Vol. II. Boca Raton, FL: CRC Press, 103141.
Zhang D, Sang T. 1999. Physical mapping of ribosomal RNA genes inpeonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization:implicationsfor phylogenyand concertedevolution.AmericanJournalof Botany 86: 735740.
276 Marcon et al. Cytogenetic Variability in Species of Selaginella