USA Trevigen Catalog 2011 1-24:Layout 1 1/24/11 5:16 PM Page 1 · BEHAVIORS AND THE TOOLS REQUIRED...
Transcript of USA Trevigen Catalog 2011 1-24:Layout 1 1/24/11 5:16 PM Page 1 · BEHAVIORS AND THE TOOLS REQUIRED...
-
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:16 PM Page 1
-
DNA DAMAGE CAUSED
BY NORMAL METABOLIC
ACTIVITIES AND ENVI-
RONMENTAL FACTORS TRIGGERS THE
DNA DAMAGE RESPONSE SYSTEM TO
ACTIVATE A DNA REPAIR PATHWAY, OR IN
THE CASE OF IRREPARABLE DAMAGE,
INDUCES APOPTOSIS.
MUTATIONS IN THE
GENES THAT ENCODE
DNA DAMAGE RESPONSE PROTEINS
MAY RESULT IN GENOMIC INSTABILITY.
PROGRESSIVE GENOMIC INSTABILITY IS
A HALLMARK OF CANCER
PROGRESSION AND MU-
TATION IN DNA DAMAGE
RESPONSE PROTEINS AND MAY BE A GEN-
ERAL FEATURE OF CANCER. EPIGENETIC
SILENCING OF GENES HAS ALSO BEEN
IMPLICATED IN THE PATHOGENESIS OF
CANCER. CANCER CELL
BEHAVIORS AND THE
TOOLS REQUIRED TO
STUDY THEIR MANY FACETS, INCLUDING
CELL MIGRATION, CELL INVASION, CELL
ADHESION AND CELL DIFFERENTIATION
ARE ALL A FOCUS OF
T R E V I G E N I N C .
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 2
-
H O W T O O R D E R
Please use product names and catalog numbers when ordering. If ordering from a quotation, please include quotenumber and the lot number for reserved lot inventory. Please provide your name, purchase order number, shippingaddress, billing address, date, and telephone number on each order.
Trevigen accepts VISA, MASTERCARD, and AMERICAN EXPRESS.
Fax, phone, e-mail, or mail your order to:
Trevigen, Inc.8405 Helgerman CourtGaithersburg, MD 208771-800-873-8443Local Phone: 301-216-2800 Fax: 301-560-4973 E-mail: [email protected]
Order online at www.trevigen.com.
International Distributors – see inside back cover.
S H I P M E N T S : All shipments are made through our carrier unless otherwise requested. Shipping charges areprepaid and added to your invoice. For RUSH orders, please advise the Trevigen’s Customer Service Representativeat the time of order.
T E R M S : Payment should be made to Trevigen, Inc., P.O. Box 7328, Gaithersburg, MD 20898, within thirty (30)days of shipment, in U.S. dollars.
C O N D I T I O N S : Trevigen products are sold for Research Use Only.
P R O D U C T U S A G E : Products manufactured and sold by Trevigen are for the customer’s research purposesonly. Resale of Trevigen products requires the express written consent of Trevigen. Trevigen products have not beenapproved for use in any clinical, diagnostic, or therapeutic applications. Obtaining license or approval to use Trevigenproducts in proprietary applications or in any non-research (clinical) applications is the customer’s exclusiveresponsibility. Trevigen will not be responsible or liable for any losses, costs, expenses, or liability arising outof the unauthorized or unlicensed use of Trevigen products.
W A R R A N T Y: Trevigen warrants that products will perform according to specifications accompanying each product.Products found not meeting specifications will be replaced, free of charge. Trevigen’s liability is limited to replacementof the product. Notice for replacement must be given within 90 days of receipt of product. Shipping discrepanciesmust be reported within 5 business days of receipt of product. All other warranties are hereby expresslydisclaimed, including but not limited to, the implied warranties of merchantability and fitness for a particular purpose,and any warranty that the products, or the use of products, manufactured by Trevigen will not infringe the patentsof one or more third parties. Trevigen shall not be liable for any consequential damages. Discrepancies with itemsordered through a Trevigen distributor must be handled with the distributor.
O T H E R : Prices are subject to change. Trevigen reserves the right to change product specifications. Products mayvary somewhat from the catalog description.
T R A D E M A R K S : CometAssay, CultreCoat, Cultrex, PathClear, TACS, TACS•XL, and Trevigen, are registeredtrademarks, and 3-D Matrix, Apoptosis Grade, CardioTACS, CometSlide, Cytonin, Cytonin IHC, DePsipher,DermaTACS, DIVAA, FLARE, FlowTACS, MitoShift, NeuroPore, NeuroTACS, PeroxyGlow, REC, TACS Blue Label,TACS-Nuclease, TACS-Sapphire, TiterTACS, TreviGel, TumorTACS, and VasoTACS are trademarks of Trevigen, Inc.
EpiDerm is a trademark of MatTek Corporation, Ashland, MA.SYBR Green I is a registered trademark of Molecular Probes, Inc., Eugene, OR.Tween 20 is a registered trademark of ICI Americas, Inc., Wilmington, DE.The PCR process is protected by patents owned by Hoffmann-LaRoche.
©2011 Trevigen, Inc. All rights reserved.
1-800-873-8443 • www.trevigen.com i
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 3
-
Overview
O V E R V I E W
1-800-873-8443 • www.trevigen.com
Dear Researcher,
Our 2011/2012 catalog reflects Trevigen’s mission: “To provide cancer researchers with the highest quality products andservices for ultimate benefit toward the prevention and cure of cancer.” Thus, we have developed products that focus onkey causative aspects emanating from DNA damage, DNA repair, and genomic instability, as well as products to detect andquantitate the downstream impact on cancer cell behavior.
Over the past few years, our specialized products for the characterization of cancer cell behavior have found numerous researchapplications in the areas of:
• Screening Therapeutics for Cancer Treatments
• Conducting Pharmacodynamic Assays on Lead Compounds
• Tumor Explant Work
• Cell Adhesion
• Metastasis
• Angiogenesis
• 3-D Structure Studies
• Apoptosis Detection
Acknowledging the complexity inherent in cancer cells and therefore cancer cell assays, Trevigen maintains a technical supportteam to assist you, the researcher, in assay selection, set up, troubleshooting as well as the interpretation of results.
We take pride in the quality of our products, which is matched by the fast turnaround and thoughtful responses of both thetechnical support and customer service teams.Our goal is to make sure you know you are able to “count on us”, at each stepof the research process, from the planning stages of your project, to data analysis/interpretation.
We organized our new catalog with our customers in mind; to make it easier for you to locate, find specifications, and toorder our products.
We also strive to keep our website up-to-date. As new products are released and new applications information becomes available for old products, we make sure the information is available immediately on our website.
Please do not hesitate to contact us at 800-873-8443 or [email protected] if you have any questions at all.
On behalf of all of us at Trevigen, Inc., I wish you continued success in your research.
Sincerely,
Michael T. Elliott
President, Trevigen, Inc.
ii
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 4
-
How To Order . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .iOverview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .ii
DNA DAMAGE AND GENOMIC INSTABILITY• Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .4
DNA Repair Gene Knockdown Cell Lines . . . . . . . . . . . .5PARP/PAR/PARG
• PARP Selection Guide . . . . . . . . . . . . . . . . . . . . . . . . . .7• PARP and PARG Selection Guide . . . . . . . . . . . . . . . . .8• Universal PARP Assay Kits . . . . . . . . . . . . . . . . . . . . . .9• PARP/Apoptosis Assay Kits . . . . . . . . . . . . . . . . . . . .10• Fluorescent Homogeneous PARP Inhibition Assay . . .11• PARP In Vivo Pharmacodynamic Assay II . . . . . . . . . .12• Universal PARG Assay Kits . . . . . . . . . . . . . . . . . . . . .14• PARP Enzyme . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .15• PARG Enzyme . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .16• PAR Polymer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .17• PARP Monoclonal Antibody . . . . . . . . . . . . . . . . . . . .18• PAR Monoclonal Antibody . . . . . . . . . . . . . . . . . . . . .19• PAR Polyclonal Antibody . . . . . . . . . . . . . . . . . . . . . . .20• Biotin‐NAD⁺ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .21• Fluorescein‐NAD⁺ . . . . . . . . . . . . . . . . . . . . . . . . . . . .22• Cell Permeabilization Solution . . . . . . . . . . . . . . . . . . .22• PARP and PARG Inhibitors . . . . . . . . . . . . . . . . . . . . .23
CometAssay™• Reagent Kits for Single Cell Gel Electrophoresis Assay .24• Silver Staining Kit . . . . . . . . . . . . . . . . . . . . . . . . . . . .26• Electrophoresis System . . . . . . . . . . . . . . . . . . . . . . .27• Control Cells . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .29• Slides and Rack System . . . . . . . . . . . . . . . . . . . . . . .30• FLARE™ Kits and Slides/ Selection Guide . . . . . . . . . .31• Fpg Assay Kit and Module . . . . . . . . . . . . . . . . . . . . . .32• hOGGI Assay Kit and Module . . . . . . . . . . . . . . . . . . .33• Endonuclease III Assay Kit and Module . . . . . . . . . . .34• UVDE Assay Kit and Module . . . . . . . . . . . . . . . . . . . .35• T4‐PDG Assay Kit and Module . . . . . . . . . . . . . . . . . .36• cv‐PDG Assay Kit and Module . . . . . . . . . . . . . . . . . . .37
DNA Damage Antibodies• Selection Guide . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .38• γ‐H2AX . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .39• 8‐oxo‐dG . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .40• FEN-1 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .41• BPDE . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .42• UVssDNA . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .43
DNA Repair Enzymes• Selection Guide . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .44• hAPE . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .45• Human DNA Polymerase β kit . . . . . . . . . . . . . . . . . . .46
• Human FEN‐1 (Flap Endonuclease) . . . . . . . . . . . . . . .47• hOGG1 DNA Glycosylase . . . . . . . . . . . . . . . . . . . . . . .48• E. Coli Endonuclease III . . . . . . . . . . . . . . . . . . . . . . .49• E. Coli Fpg DNA Glycosylase . . . . . . . . . . . . . . . . . . . .50• S. pombe UVDE . . . . . . . . . . . . . . . . . . . . . . . . . . . . .51• T4 Endonuclease V . . . . . . . . . . . . . . . . . . . . . . . . . . .52• Cv‐PDG . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .54• E. Coli Endonuclease IV . . . . . . . . . . . . . . . . . . . . . . .56• UNGase DNA Glycosylase . . . . . . . . . . . . . . . . . . . . . .58• Aag DNA Glycosylase . . . . . . . . . . . . . . . . . . . . . . . . .60• Mug DNA Glycosylase . . . . . . . . . . . . . . . . . . . . . . . . .61• MutY DNA Glycosylase . . . . . . . . . . . . . . . . . . . . . . . .62• TDG DNA Glycosylase . . . . . . . . . . . . . . . . . . . . . . . . .63• Dpo4 Polymerase . . . . . . . . . . . . . . . . . . . . . . . . . . . .64• Human Ku 70/80 Complex . . . . . . . . . . . . . . . . . . . . .65
CANCER CELL BEHAVIOR• Overview/ Cultrex® Product Selection Guide . . . . . . . .68
Basement Membrane Extract (BME), Mouse . . . . . . . .70• BME Selection Guide . . . . . . . . . . . . . . . . . . . . . . . . . .71• BME PathClear® . . . . . . . . . . . . . . . . . . . . . . . . . . . . .72• BME High Protein Concentration . . . . . . . . . . . . . . . . .74• BME Stem Cell Qualified . . . . . . . . . . . . . . . . . . . . . . .75• BME 3‐D Culture Matrix™ . . . . . . . . . . . . . . . . . . . . . .76
ECM Component Proteins and Reagents• Overview/ Selection Guide . . . . . . . . . . . . . . . . . . . . .77• Human BME . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .80• Human Fibronectin . . . . . . . . . . . . . . . . . . . . . . . . . . .81• Human Vitronectin . . . . . . . . . . . . . . . . . . . . . . . . . . .82• Mouse Laminin I, . . . . . . . . . . . . . . . . . . . . . . . . . . . .83• Mouse Laminin I PathClear® . . . . . . . . . . . . . . . . . . . .84• Rat Collagen I . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .85• Mouse Collagen IV . . . . . . . . . . . . . . . . . . . . . . . . . . .86• Bovine Collagen I . . . . . . . . . . . . . . . . . . . . . . . . . . . .87• Bovine Fibronectin . . . . . . . . . . . . . . . . . . . . . . . . . . .88• Bovine Vitronectin . . . . . . . . . . . . . . . . . . . . . . . . . . . .89• Poly‐D‐Lysine . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .90• Poly‐L‐Lysine . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .91• 3‐D Culture Matrix™ Mouse Laminin . . . . . . . . . . . . .92• 3‐D Culture Matrix™ Rat Collagen I . . . . . . . . . . . . . . .93
Cancer Cell Assays• Overview/ Selection Guide . . . . . . . . . . . . . . . . . . . . .94• Directed In Vivo Angiogenesis Assay (DIVAA™) . . . . .95• In Vitro Angiogenesis Assay Tube Formation Kit . . . . .97• In Vitro Angiogenesis Assay Endothelial Cell
Invasion Kit . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .98• CultreCoat® Vascular Permeability Assays . . . . . . . . .99• Cell Invasion Assays . . . . . . . . . . . . . . . . . . . . . . . . .100
1-800-873-8443 • www.trevigen.com
Table of Contents
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 5
-
1-800-873-8443 • www.trevigen.com
• Cell Invasion Optimization System . . . . . . . . . . . . . .100• Cell Migration Assays . . . . . . . . . . . . . . . . . . . . . . . .100• CultreCoat® Cell Adhesion Assays . . . . . . . . . . . . . . .102• Cell Staining Kit . . . . . . . . . . . . . . . . . . . . . . . . . . . . .104
3‐D Products• 3‐D Culture Matrix™ BME . . . . . . . . . . . . . . . . . . . . .105• 3‐D Culture Matrix™ Mouse Laminin I . . . . . . . . . . .106• 3‐D Culture Matrix™ Rat Tail Collagen I . . . . . . . . . .107• 3‐D Proliferation Assays . . . . . . . . . . . . . . . . . . . . . .108• 3‐D Culture Cell Harvesting Kit . . . . . . . . . . . . . . . . .109• MTT/XTT Cell Proliferation Assay . . . . . . . . . . . . . . .111• Calcein‐AM Cell Viability Assay . . . . . . . . . . . . . . . . .111
APOPTOSIS DETECTION• Overview/ Selection Guide . . . . . . . . . . . . . . . . . . . .114
Annexin V Assays• TACS® Annexin V‐FITC Kit . . . . . . . . . . . . . . . . . . . . .118• TACS® Annexin V‐Biotin Kit . . . . . . . . . . . . . . . . . . . .120
DNA Fragmentation• TACS.XL® In Situ Apoptosis Detection Kits . . . . . . . .121• TACS® 2TdT In Situ Apoptosis Detection Kits . . . . . .123• Specialty In Situ Apoptosis Detection/
Selection Guide . . . . . . . . . . . . . . . . . . . . . . . . . . . . .125• CardioTACS™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .126• DermaTACS™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .128• NeuroTACS™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .130• TumorTACS™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .132• VascoTACS™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .134• TiterTACS™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .135• FlowTACS™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .137• TACS® DNA Laddering Kit . . . . . . . . . . . . . . . . . . . . .139
Mitochondrial Events• DePsipher™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .140• MitoShift™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .142• PARP/Apoptosis Assay Kits . . . . . . . . . . . . . . . . . . .144
Cell Proliferation• TACS® MTT Cell Proliferation Assay . . . . . . . . . . . . .145• TACS® XTT Cell Proliferation Assay . . . . . . . . . . . . . .146• Calcein AM Cell Viability Kit . . . . . . . . . . . . . . . . . . . .147
Antibodies• Selection Guide . . . . . . . . . . . . . . . . . . . . . . . . . . . . .148• Bax . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .149• Bcl‐2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .151• Bcl‐XL . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .152• PBR Antibody and Protein . . . . . . . . . . . . . . . . . . . . .153• Cleaved Caspase‐3 . . . . . . . . . . . . . . . . . . . . . . . . . .154• G3PDH . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .155
Apoptosis Research Accessories . . . . . . . . . . . . . . .156Apoptosis Product Use . . . . . . . . . . . . . . . . . . . . . .159
STEM CELL PRODUCTS• Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .164• Stem Cell Qualified BME . . . . . . . . . . . . . . . . . . . . . . . .165• Rat Mesenchymal Stem Cell Growth
and Replenishment products . . . . . . . . . . . . . . . . . . . .166• Mesenchymal Stem Cell Adipogenic
Differentiation Kit . . . . . . . . . . . . . . . . . . . . . . . . . . . . .167• Mesenchymal Stem Cell Osteogenic
Differentiation Kit . . . . . . . . . . . . . . . . . . . . . . . . . . . . .168
OXIDATIVE STRESS• Overview/ Selection Guide . . . . . . . . . . . . . . . . . . . . . .170• 8‐oxo‐dG ELISA Kit . . . . . . . . . . . . . . . . . . . . . . . . . . . .171• 8‐oxo‐dG Monoclonal Antibody . . . . . . . . . . . . . . . . . .172• HT Superoxide Dismutase Assay Kit . . . . . . . . . . . . . . .173• Superoxide Dismutase Assay Kit . . . . . . . . . . . . . . . . . .174• HT Glutathione Assay Kit . . . . . . . . . . . . . . . . . . . . . . . .175• HT Glutathione Peroxide Assay Kit . . . . . . . . . . . . . . . .176• HT Glutathione Reductase Assay Kit . . . . . . . . . . . . . . .177• Glutathione Reductase Assay Kit . . . . . . . . . . . . . . . . . .178
MOLECULAR BIOLOGY REAGENTS• Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .180• Genomic DNA Isolation Kit . . . . . . . . . . . . . . . . . . . . . .181• TreviGel™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .182• Orange G Loading Dye . . . . . . . . . . . . . . . . . . . . . . . . .183• Electrophoresis Buffer . . . . . . . . . . . . . . . . . . . . . . . . . .183• PBS . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .183• TdT DNA Polymerase . . . . . . . . . . . . . . . . . . . . . . . . . .184• Calf Thymus DNA, Herring Sperm DNA,• Salmon Sperm DNA . . . . . . . . . . . . . . . . . . . . . . . . . . .184• Proteinase K . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .184
TREVIGEN CELL ASSAYS (TCA) SERVICES• CometAssay™ . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .186• PARP Assay . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .186• PARG Assay . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .186• Pharmacodynamic PARP Assay . . . . . . . . . . . . . . . . . .186• Cell Invasion Assay . . . . . . . . . . . . . . . . . . . . . . . . . . . .186• Tube Formation Assay . . . . . . . . . . . . . . . . . . . . . . . . . .186
Price List . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .187Appendix . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .192Index . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .193Technical Resources . . . . . . . . . . . . . . . . . . . . . . . .195International Distributors . . . . . . . . . . . . . . . . . . . . .198
Table of Contents
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 6
-
Cancer Cell Behavior
Stem Cell Products
Apoptosis Detection
Oxidative Stress
DNA DAMAGE AND GENOMIC INSTABILITY
Molecular Biology Reagents
DN
A D
AM
AG
E A
ND
GE
NO
MIC
IN
STA
BIL
ITY
DNA DAMAGE AND GENOM
IC INSTABILITY
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 7
-
1-800-873-8443 • www.trevigen.com
Trevigen is pleased to provide highly qualified enzymes, antibodies, and assay kits for DNAdamage and repair research. In this catalog, we have NEW and UNIQUE additions to our productline to complement and provide your research efforts with flexibility. As always, our productsare accompanied by detailed product data sheets or instructions for use backed with experttechnical support.
D N A R E PA I R D E F I C I E N T C E L L L I N E SDNA repair pathways maintain the integrity of the genome reducing the onset of cancer, disease and aging phenotypes. However, the requirement for DNA repair and genomemaintenance in response to radiation and genotoxic chemotherapies implicates DNA repairproteins as prime targets for improving response to currently available anti-cancer approaches. To further study repair pathways, Trevigen offers panels of human cell lineseach deficient in proteins associated with DNA repair. The first panel consists of cell linesdeficient for proteins in the Base Excision Repair Pathway. For the availability of other DNArepair deficient cell lines contact Trevigen.
PA R P / PA R / PA R GPARP-1 contributes to the sequence of events that occurs during DNA base excision repair.Poly (ADP-ribose) polymerase (PARP) catalyzes the NAD+ - dependant addition of poly(ADP-ribose) (PAR) onto itself and adjacent nuclear proteins in response to DNA damage.
Since PAR is often highly branched and degraded by poly(ADP-ribose) glycohydrolase(PARG), Trevigen offers kits that measure the in vivo and in vitro activities of PARP andPARG. One hallmark of apoptosis is caspase-mediated cleavage and inactivation of PARP-1.The PARP Apoptosis Kits measure decreasing levels of PARP activity as cells move throughthe apoptosis pathway. The Homogeneous PARP Inhibition Assay is well suited for the largescale screening of compound libraries for candidate PARP inhibitors. Our Universal PARPand PARG Assays are designed to analyze small numbers of inhibitors and recommendedfor providing accurate IC50 information. Inhibitors to PARP can be characterized by in vitro assays. Their in vivo activity in cell or tissue extracts, or peripheral blood mononuclear cellscan be quantitatively measured using Trevigen’s validated PARP in vivo PharmacodynamicAssay. Biotinylated NAD+, PAR and PARP antibodies, key reagents for the described assays, are also available as individual components.
C O M E TA S S AY ®The single cell gel electrophoresis or CometAssay® is a state-of-the-art technique forquantitating DNA damage and repair from in vivo and in vitro samples of eukaryotic cellsand some prokaryotic cells. In conjunction with Trevigen’s new CometAssay® ElectrophoresisSystem, which eliminates known causes of assay variability, it is the only technique that directly measures DNA damage in individual cells and as a result has rapidly gained importance in the fields of genetic toxicology and human biomonitoring. Trevigen’sCometAssay® measures double strand breaks (DSBs), single strand breaks (SSBs), alkalilabile sites, oxidative DNA base damage, DNA-DNA/ DNA-protein/DNA-Drug crosslinkingand DNA repair.
D N A R E PA I R E N Z Y M E S A N D A N T I B O D I E STrevigen’s unique FLARE™ Assays characterize DNA damage in single cells using a varietyof DNA repair enzymes that recognize oxidative and UV damage in conjunction withTrevigen’s CometAssay® Electrophoresis System. A variety of DNA repair enzymes andantibodies are available as individual components with optimized assay conditions andbuffers for research in areas of double-strand break repair, base excision repair, DNAmethylation, oxidative damage, UV damage, and radiation damage.
Please check our website for the latest product additions and product information.
Overview
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T Y DN
A DA
MAG
E AN
D GE
NOM
IC IN
STAB
ILIT
YOV
ERVI
EW
4
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 8
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
KNOCKDOWN CELL LINES
5
DNA repair proteins maintain the stability of the genome. When repair protein function isimpaired through mutations the genome may become unstable, which is a hallmark of solidtumors. The availability of this panel of knockdown cell lines will permit scientists to studythe molecular etiology of tumor genomic instability and to exploit it in oncology research.The first panel covers the Base Excision Repair pathway.
Trevigen’s Base Excision Repair Knockdown Cell Lines are target specific LN428 (glioblas-toma) shRNA lentivirus transduced cells. They are rigorously qualified and mycoplasmafree. The percent knockdown levels range from 63% -98% as evaluated by RT-PCR.Lentiviruses are maintained by puromycin selection.
F E AT U R E S :Tested negative for Mycoplasma
Tested negative for: Human immunodeficiency virus (HIV1, HIV2), Hepatitis viruses (A, B,and C), Human T-lymphotropic virus (HTLV 1, HTLV 2), Epstein Barr, Hantaviruses Hantaan(Seoul, Sin, Nombre), Herpes simplex (1+2,) Human cytomegalovirus, Human herpes virus(6+8), Human adenovirus, Varicella virus and Lymphocytic Choriomeningitis virus.
O R D E R I N G I N F O R M AT I O N :
* Cell lines are provided under an MTA. Please inquire at [email protected]
DNA Repair Gene Knockdown Cell LinesTools to Study Genomic Instability and Genotoxic StressDNA Repair Gene Knockdown Cell Lines*Base Excision Repair
Target Knockdown cell line Catalog number # of cells Size Price R
APE1 KD-BER¹-LN428³-APE1 5517-001-01 1x106 1 vial $895 90%APE2 KD-BER-LN428-APE2 5518-001-01 1x106 1 vial $895 80%BRCA1* KD-HR²-LN428-BRCA1 5502-001-01 1x106 1 vial $895 83%XRCC1 KD-BER-LN428-XRCC1 5516-001-01 1x106 1 vial $895 81%MBD4 KD-BER-LN428-MBD4 5506-001-01 1x106 1 vial $895 72%MPG KD-BER-LN428-MPG 5511-001-01 1x106 1 vial $895 98%MutYH KD-BER-LN428-MutYH 5512-001-01 1x106 1 vial $895 87%NEIL1 KD-BER-LN428-NEIL1 5513-001-01 1x106 1 vial $895 92%NEIL2 KD-BER-LN428-NEIL2 5507-001-01 1x106 1 vial $895 86%NEIL3 KD-BER-LN428-NEIL3 5508-001-01 1x106 1 vial $895 95%NTHL1 KD-BER-LN428-NTHL1 5505-001-01 1x106 1 vial $895 91%OGG1 KD-BER-LN428-OGG1 5504-001-01 1x106 1 vial $895 63%PARP1 KD-BER-LN428-PARP1 5500-001-01 1x106 1 vial $895 72%PARP2 KD-BER-LN428-PARP2 5514-001-01 1x106 1 vial $895 83%PARP3 KD-BER-LN428-PARP3 5515-001-01 1x106 1 vial $895 70%PARG KD-BER-LN428-PARG 5501-001-01 1x106 1 vial $895 84%SMUG1 KD-BER-LN428-SMUG1 5510-001-01 1x106 1 vial $895 63%TDG KD-BER-LN428-TDG 5519-001-01 1x106 1 vial $895 74%UNG KD-BER-LN428-UNG 5509-001-01 1x106 1 vial $895 87%Control KD-BER-LN428-Control 5503-001-01 1x106 1 vial $475 N/A
# of cellsper vial
%KD byRT-PCR
¹BER-Base Excision Repair ²HR- Homologous Recombination³LN428- Human malignant glioblastoma cell line*BRCA1 participates in the Homologous Recombination Pathway and demonstrates synthetic lethality in combination with PARP1.
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 9
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T Y KN
OCKD
OWN
CELL
LIN
ES
6
All Trevigen knockdown cell lines areevaluated by RT-PCR and Western blotswhen reliable immunoreagents are available. Additional functional assaysare performed when feasible.
Enzymatic Assay for
MPG activity
Lane 1: Buffer alone
Lane 2: Control extract
Lane 3: MPG knockdown
Western Blot of MPG protein
Lane 1: Molecular Weight Markers
Lane 2: Scramble RNA Control
Lane 3: MPG knockdown extract
Lane 4: Control cell line
DNA Repair Gene Knockdown Cell LinesBRCA1 Knockdown cell line is sensitive to PARP1 inhibitors
R E F E R E N C E S :1. Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T: DNA Repairand Mutagenesis, 2nd Edition. Washington, D.C.: ASM Press; 2006.2. Alberts B: Redefining cancer research. Science 2009, 325(5946):1319(PMID:19745119).3. Ljungman M: Targeting the DNA damage response in cancer. Chem Rev 2009,109(7):2929-2950 (PMID:19545147).4. Peralta-Leal A, Rodriguez MI, Oliver FJ: Poly(ADP-ribose)polymerase-1 (PARP-1) incarcinogenesis: potential role of PARP inhibitors in cancer treatment. Clin Transl Oncol2008, 10(6):318-323 (PMID:18558578).
S T O R A G E :Liquid Nitrogen
L I M I T E D U S E N O T I C E :Trevigen’s DNA Repair Gene Knock Down Cell Lines can only be obtained under a MaterialsTransfer Agreement executed with Trevigen, Inc. The Materials Transfer Agreement conveysto the buyer the non-transferable right to use the purchased amount of the product and allreplicates and derivatives for research purposes conducted by the buyer, in their laboratoryonly. For details on the Materials Transfer Agreement or for information on purchasing alicense for these products for purposes other than research, contact Trevigen, Inc.
Knockdown cell line growth in presence of PARP inhibitor.
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 10
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
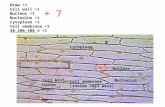
PARP ASSAY SELECTION GUIDE
7
PARP/ PAR/ PARGPARP Assay Selection Guidehibitors
poly-ADP ribose
EtOH
nicotinamide
Activated +DNA
PARP 1active
PARP 1inactive
Acetaldehyde
ResazurinFluorescent Resorufin
[NAD+]
Step 1: PARP Reaction
Step 2: Cycling Assay
NADH +H
AD
PARP Homogeneous Assay
Diaphorase
HRP
ChemiluminescentDetection
Colorimetric orChemiluminescent
Detection
Goat anti-rabbit IgG-HRP
Anti-PAR Polyclonal
Anti-PAR MonoclonalImmobilized
PAR
HRP
Streptavidin
Biotin
PAR
Histones
Evaluation of Inhibitor BehaviorIn Vivo or in Cultured Cells
Verification of Lead Compounds orLimited Inhibitor Screening Campaigns.Determination of IC
50 Values
HT Universal PARP Assay Kits# 4677-096-K Colorimetric# 4676-096-K Chemiluminescent
Determine PARP Activityin Cell Lysates
Trevigen’s PARP Pharmacodynamic Assay
Trevigen’s Universal PARP Assay
HRP
Colorimetric orChemiluminescent
Detection
Goat anti-mouse IgG-HRP
Anti-PAR Monoclonal
PAR
Histones
Trevigen’s PARP Apoptosis Assay
High Throughput Screening for PARP Inhibitors
hv ~ [NAD+]remaining after Step 1
HT F Homogeneous PARPInhibition Assay Kit# 4690-096-K
HT PARP in vivoPharmacodynamic
Assay II# 4520-096-K
HT PARP/ Apoptosis Assay Kit# 4684-096-K Colorimetric# 4685-096-K Chemiluminescent
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 11
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T Y
8
PARP
/ PAR
/ PAR
G AS
SAY
SELE
CTIO
N GU
IDE
Summary of PARP and PARG Assay Details:
Catalog # Application(s) Detection Method Test Sample Sensitivity Time* Page Universal PARP Assay Kit**• Colorimetric Detection 4677-096-K • Chemiluminescence Detection 4676-096-K PARP/Apoptosis Kit**• Colorimetric Detection 4684-096-K • Chemiluminescence Detection 4685-096-K
Homogeneous Fluorescence 4690-096-K
PARP in vivo Pharmacodynamic Assay II** 4520-096-K
PARG Assay Kits**• Colorimetric Detection 4683-096-K • Chemiluminescence Detection 4682-096-K
*Actual Times may vary depending upon the number of samples being prepared and assayed.**Provided with a 96 strip well plate.
y
PARP Inhibitor ScreenIC50 Determination
PARP Inhibitor Screen PARP Activity in Lysates
HT PARPInhibitor Screen
PARP Inhibition in vivo PAR Levels in Lysates
PARG Inhibitor Screen
Biotin NAD+ Histone ribosylation 3 hr
3 hr
1.5 hr
6.5 hr
2 hr
PAR Ab Histone ribosylation
NAD+ Consumption PARP ribosylation
PAR Ab In vivo PARylation
Biotin NAD+ PAR degradation
PARP Inhibitors
PARP Inhibitors Cell Lysates
PARP Inhibitors
PMBC Lysates Tissue Lysates
PARG Inhibitors
0.01-1 Units PARP
0.1 - 10 mUnits PARP 500 Cells
10% PARP inhibition
2-1000 pg/ml PAR
250 pg PARG
PARP Inhibition Assay
9
10
11
12
14
PARP/ PAR/ PARGPARP and PARG Assay Selection Guide
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 12
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
9
UNIVERSAL PARP ASSAY
PARP/ PAR/ PARGUniversal PARP Assay Kits
Poly (ADP-ribose) polymerase (PARP) catalyzes the NAD+-d ependent addition of poly(ADP-ribose) to itself and adjacent nuclear proteins, such as histones, in response to DNAdamage. PARP contributes to the sequence of events that occurs during DNA base excisionrepair. Whereas PARP-mediated induction of necrosis can occur by extensive depletion of theintracellular NAD+ pool, the cleavage of PARP-1 promotes apoptosis by preventing DNArepair-induced survival and by blocking energy depletion-induced necrosis. Moreover, PARPinhibition promotes chemosensitization and radiosensitization of tumors. Trevigen's HTUniversal PARP Assay Kits measure the incorporation of biotinylated poly(ADP-ribose) ontohistone proteins in a 96-well strip well format. This assay is ideal for the determination of IC50values of known or suspected PARP inhibitors.
For your convenience, Trevigen provides histone-coated strip wells. This format significantlyreduces assay time.
F E AT U R E S :• Colorimetric or Chemiluminescent readout• 96 well format• Sensitive – detects 10 mU PARP/well • Assay Time ~3 hrs
A P P L I C AT I O N S :• Assay inhibitors and activators of PARP activity• Determination of IC50 values for PARP Inhibitors
K I T C O M P O N E N T S :PARP-HSAHistone-Coated Strip Wells PARP Buffer and CocktailColorimetric or Chemiluminescent Detection ReagentsActivated DNAStrep HRP and Diluent3-Aminobenzamide (known PARP inhibitor)
R E L AT E D P R O D U C T S :PARP Inhibitors (see page 23)
R E F E R E N C E S :1. Satoh MS, Lindahl T. 1992. Role of poly(ADP-ribose) formation in DNA repair.Nature 356: 356–8.2. Golstein P, Kroemer G. 2007. Cell death by necrosis: Towards a molecular definition.Trends Biochem Sci 32: 37-43.3. D’Amours D, Sallmann FR, Dixit VM, Poirer GG. 2001. Gain-of-function of poly(ADP-ribose) polymerase-1 upon cleavage by apoptotic proteases: implications forapoptosis. J Cell Science 114: 3771-78.4. Eliasson M, Sampei K, Mandir AJ. 1997. Poly(ADP-ribose) polymerase gene disruptionrenders mice resistant to cerebral ischemia. Nat Med 3:1089- 95.5. Jagtap P, Szabó C. 2005. Poly(ADP-ribose) polymerase and the therapeutic effects of itsinhibitors. Nat Rev Drug Discov. 4:421-40.6. Pieper AA, Verma A, Zhang J, Snyder SH. 1999. Poly(ADP-ribose) polymerase, nitricoxide and cell death. Trends Pharmacol Sci 20:171-81.7. Thiemermann C, Bowes J, Myint FP, Vane JR. 1997. Inhibition of the activity ofpoly(ADP-ribose) synthase reduces ischemia-reperfusion injury in the heart and skeletalmuscle. Proc Natl Acad Sci USA 94:679-83.8. Kauppinen TM, Swanson RA. 2005. Poly(ADP-ribose) polymerase-1 promotesmicroglial activation, proliferation, and matrix metalloproteinase-9-mediated neuron death.J Immunol. 174:2288-96.9. http://www.trevigen.com/MarketingFlyers/JDS_PARP2008_Poster.pdf
4667-50-06 $85 Activated DNA for PARP Assay,500 µl
4676-096-K $445 HT Universal Chemiluminescent PARPAssay Kit with Histone-coated StripWells, 96 tests
4677-096-K $445 HT Universal Colorimetric PARP AssayKit with Histone-coated Strip Wells,96 tests
4677-096-P $60 Histone-Coated Natural Strip Wells,96 tests
4678-096-P $60 Histone-Coated White Strip Wells,96 tests
Bulk quantites available; please inquire
O R D E R I N G I N F O R M A T I O N
Colorimetric orChemiluminescent
Detection
HRP
Streptavidin
Biotin
PAR
Histones
Trevigen’s Universal PARP Assay
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 13
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YPA
RP/A
POPT
OSIS
ASS
AY
10
PARP/ PAR/ PARGPARP/Apoptosis Assay Kits
During apoptosis, PARP-1 which catalyzes the NAD+-dependent addition of poly (ADP-ribose)(PAR) onto various cytoplasmic and nuclear proteins, is cleaved from about 116 kDa to85 kDa. Moreover, this enzyme is a therapeutic target for BRCA1and BRCA2 associatedbreast cancers. The HT PARP/Apoptosis Assay is ideal for measuring the activity of PARP incell extracts before and during apoptosis. This ELISA semi-quantitatively detects PAR deposited onto immobilized histone proteins in a 96-well format. An anti-PAR monoclonalantibody, goat anti-mouse IgG-HRP conjugate, and HRP substrate are used to generate a signal. Thus, signals correlate with PARP activity. Etoposide is a topoisomerase II inhibitorthat stabilizes this enzyme after it cleaves DNA. It is included as a control apoptosis inducer.
F E AT U R E S :• Colorimetric or Chemiluminescent readout• 96 well format• Highly sensitive – detects 0.1 mU PARP ~500 cells• Dynamic range between 0.1 to 10 mU PARP• Requires 10-100 ng extract for detection• Assay Time ~3 hrs
A P P L I C AT I O N S :• Measure activity in cells, primary or tumor cells• Measure activity before and after apoptosis• Measure effect of PARP inhibitors using cell extracts
K I T C O M P O N E N T S :PARP-HSA and Buffer Anti-PAR Monoclonal Antibody Activated DNA NAD+
Histone-Coated Strip Wells PAR mAb and DiluentHRP Conjugate Etoposide Colorimetric or Chemiluminescent Detection Reagents
R E F E R E N C E S :1. Lawen A. 2003. Apoptosis—an introduction. BioEssays 25:888-896.2. Okada H, Mak TW. 2004. Pathways of apoptotic and non-apoptotic death in tumor cells.Nat Rev Cancer 4:592-603.3. Miller MS, Zobre C, Lewis M. 1993. In vitro neuroprotective activity of inhibitors ofpoly-ADP ribose polymerase. Soc Neurosci Abstr 19.16564. Piper AA, Verma A, Zhang J, Snyder SH. 1999. Poly(ADP-ribose) polymerase, nitricoxide and cell death. Trends in Pharmacological Sciences 20:171-181.5. Thiemermann C, Bowes J, Myint FP, and Vane JR. 1997. Inhibition of the activity ofpoly(ADP-ribose) synthase reduces ischemia-reperfusion injury in the heart and skeletalmuscle. Proc Natl Acad Sci USA 94:679-683.6. Virag L, Szabo C. 2002. The therapeutic potential of Poly(ADP-Ribose) Polymeraseinhibitors. Pharmacological Reviews 54:375-429.7. Baldwin EL, Osherhoff N. 2005. Etoposide, Topoisomerase II and Cancer. Curr. Med.Chem. Anti-Cancer Agents 5:363-372.8. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M,Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors ofpoly(ADP-ribose) polymerase. Nature 434:913-7.9. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M,Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. 2005.Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature434:917-21.10. Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H,Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. 2009.Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers.N Engl J Med 361:123-34.
4684-096-K $495 HT Universal ColorimetricPARP/Apoptosis Assay Kit, 96 tests
4685-096-K $495 HT Universal ChemiluminescentPARP/Apoptosis Assay Kit, 96 tests
O R D E R I N G I N F O R M A T I O N
ASSAY DESIGN:
Step 1: Ribosylation of histones by PARP.
• PAR polymer synthesized onto immobilized histonesusing PARP-HSA or cell extracts
• PARP inhibitors and caspase 3 cleavage preventsynthesis of PAR polymer.
Step 2: Detection to measure incorporation of PARattached to histones via PARP monoclonal antibody. • Color/light output (Signal) is proportional to
PARP activity.• Monitor apoptosis by a decrease in PARP Signal.
HRP
Colorimetric orChemiluminescent
Detection
Goat anti-mouse IgG-HRP
Anti-PAR Monoclonal
PAR
Histones
Trevigen’s PARP Apoptosis Assay
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:17 PM Page 14
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
PARP INHIBITION ASSAY
11
PARP/ PAR/ PARGHT Fluorescent Homogeneous PARP Inhibition Assay
Trevigen's HT Fluorescent Homogeneous PARP Inhibition Assay is a highly sensitivefluorescent screening assay for the rapid identification of PARP-1 inhibitors in an in vitrosystem. This one hour endpoint assay is performed in two successive steps requiring onlythe consecutive addition of reaction components. A PARP autoribosylation reaction is firstperformed, followed by a detection step. The remaining NAD+ level is determined in acycling reaction involving alcohol dehydrogenase and diaphorase. Each time NAD+ cyclesthrough these coupled reactions, a molecule of highly fluorescent resorufin is generated.The assay can be used to determine relative IC50 values for PARP inhibitors and is capable ofdetecting as little as 10% inhibition of PARP-1 activity. Inhibitors are identified by an increasein fluorescent signal when PARP mediated NAD+ depletion is inhibited.
F E AT U R E S :• Highly sensitive fluorescent assay• Detect as little as 10% inhibition of PARP Activity• Inhibitors identified by increase in Fluorescent Signal• No wash steps• 96 test size• Rapid – Assay takes ~ 1.5 hr
A P P L I C AT I O N S :• Ideal for screening compounds which potentially inhibit PARP activity• Determination of relative IC50 values for PARP Inhibitors
K I T C O M P O N E N T S :PARP-HSA and BufferActivated DNANAD+
10X Cycling Enzymes10X ResazurinStop Solution
R E F E R E N C E S :1. Meyer-Ficca, M.L., et al., 2005. Poly(ADP-ribose) polymerases: managing genomestability. Int J Biochem Cell Biol, 37:920-6.2. Hassa, P.O., et al., 2006. Nuclear ADP-ribosylation reactions in mammalian cells: where arewe today and where are we going? Microbiol Mol Biol Rev, 70:789-829.3. Virag, L. and C. Szabo, 2002. The therapeutic potential of poly(ADP-ribose) polymeraseinhibitors. Pharmacol Rev 54:375-429.4. Tutt, A.N., et al., 2005. Exploiting the DNA repair defect in BRCA mutant cells in the designof new therapeutic strategies for cancer. Cold Spring Harb Symp Quant Biol, 70:139-48.5. Bryant, H.E., et al., 2005. Specific killing of BRCA2-deficient tumours with inhibitors ofpoly(ADP-ribose) polymerase. Nature 434:913-7.6. Ratnam, K. and J.A. Low 2007. Current development of clinical inhibitors ofpoly(ADP-ribose) polymerase in oncology. Clin Cancer Res 13:1383-8.
ASSAY DESIGN:
Step 1: PARP reaction requiring NAD+ is performed.
• As PAR polymer is synthesized NAD+ is consumed from the reaction mixture.
• PARP inhibitors prevent NAD+ consumption.Step 2: Cycling Assay is performed to quantify
NAD+ remaining from Step 1. • Each time NAD+ cycles, a molecule of highly
fluorescent resorufin is generated. • PARP inhibitors are identified by an increase
in fluorescent Signal.
poly-ADP ribose
EtOH
nicotinamide
Activated +DNA
PARP 1active
PARP 1inactive
Acetaldehyde
ResazurinFluorescent Resorufin
[NAD+]
Step 1: PARP Reaction
Step 2: Cycling Assay
NADH +H
AD
PARP Homogeneous Assay
Diaphorase
hv ~ [NAD+]remaining after Step 1
4690-096-K $450 HT Fluorescent Homogeneous PARPInhibition Assay, 96 tests
O R D E R I N G I N F O R M A T I O N
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 15
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YPA
RP IN
VIV
OPH
ARM
ACOD
YNAM
IC A
SSAY
12
PARP catalyzes the NAD+-dependent addition of poly (ADP-ribose) (PAR) onto itself andadjacent nuclear proteins. This enzyme is a therapeutic target for BRCA1 and BRCA2 associated breast cancers. To address the need to monitor PARP activity among different individuals and within cells, Trevigen's improved and validated HT PARP in vivo Pharmacody-namic Assay II measures net PAR levels in tissue or cellular extracts and has been used todocument differences in PAR levels among tumor lysates, organs and xenografts. The HTPARP in vivo Pharmacodynamic Assay II employs a 96 well plate, pre-coated with Trevigen’smonoclonal PAR antibody as the capture agent, and anti-PAR polyclonal rabbit antibody asthe detecting agent.
F E AT U R E S :• Pre-coated 96 well capture antibody plates• High signal to noise ratio• Detection sensitivity of 2 pg/ml of PAR• Broad linear dynamic range to 1,000 pg/ml• Reduced inter-assay variability• Validated assay that measures drug action on PARP in both in vivo and in vitro settings• 96 test size
A P P L I C AT I O N S :• Quantitation of PAR in peripheral blood mononuclear cells, tissue culture cells, and tumor lysates from different tissues, organs and xenografts.• Monitoring the efficacy of PARP inhibitors on PAR formation in vivo. • Verifying observations of enhanced cancer cell cytotoxicity arising from PARP inhibitor/ anticancer drug combination therapy.• Facilitating development of PARP and PARG targeted therapeutics.
K I T C O M P O N E N T S :PAR Standard and Sample Buffer PAR Polyclonal Detecting Ab and DiluentGoat anti-Rabbit IgG-HRP Detection ReagentsCell Lysis Reagent and 20% SDS DNase I and Mg CationJurkat Cell Lysate Standard Controls (Low, Medium, and High)Pre-coated 96-stripwell plate and sealers
R E F E R E N C E S :1. Virag, L., Szabo, C. 2002. The therapeutic potential of Poly(ADP-Ribose) Polymeraseinhibitors. Pharmacological Reviews 54:375-429.2. Curtin NJ. 2005. PARP inhibitors for cancer therapy. Expert Rev Mol Med 7:1-20.3. Kinders JK, Hollingshead M, Khin S, Rubinstein L, Tomaszewski JE, Doroshow JH,Parchment RE, and the National Cancer Institute Phase 0 Clinical Trials Team. 2008.Preclinical Modeling of a Phase 0 Clinical Trial: Qualification of a Pharmacodynamic Assayof Poly (ADP-Ribose) Polymerase in Tumor Biopsies of Mouse Xenografts.Clin Cancer Res 14:6877-85.4. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M,Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors ofpoly(ADP-ribose) polymerase. Nature 434:913-7.5. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M,Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. 2005.Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy.Nature 434:917-21.6. Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H,Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. 2009.Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers.N Engl J Med 361:123-34.
PARP/ PAR/ PARGHT PARP in vivo Pharmacodynamic Assay II
4520-096-K $1,250HT PARP in vivo PharmacodynamicAssay II, 96 tests
O R D E R I N G I N F O R M A T I O N
HRP
ChemiluminescentDetection
Goat anti-rabbit IgG-HRP
Anti-PAR Polyclonal
Anti-PAR MonoclonalImmobilized
PAR
Trevigen’s PARP Pharmacodynamic Assay
ASSAY DESIGN:
Step 1: Immobilized PAR mAb captures cellular PARand PAR attached to proteins in prepared lysates.
Step 2: Binding of PAR polyclonal detecting Ab tocaptured PAR.
Step 3: Measure captured PAR via binding of goatanti-rabbit IgG-HRP with chemiluminescent detection.
• Light output (Signal) correlates with the amount ofcellular PAR.• Allows quantification of PAR in biopsies of tumorxenografts, tissue samples, peripheral bloodmononuclear cells and tissue culture cells.
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 16
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
PARP IN VIVOPHARM
ACODYNAMIC ASSAY
13
PARP/ PAR/ PARGHT PARP in vivo Pharmacodynamic Assay II
A S S AY VA L I D AT I O N :
PAR level measured in Jurkat, PBMC and Xenograft cell lysate
0.0
200.0
400.0
600.0
800.0
1000.0
Jurkat (High) Jurkat(Medium)
Jurkat (Low) PBMC Xeno
PA
R (
pg
/ml)
Assay Validation Using Control Lysates
PAR levels in three donors' PBMC lysates
0.0
20.0
40.0
60.0
80.0
100.0
120.0
J1 J2 J3 K1 K2 K3 L1 L2 L3
PA
R (p
g/m
l)
Assay Validation Using Donor Cells. Cells obtained from three different donors were lysed and assayed on three different days.The means and standard deviations of each determination are shown.
PAR Standard Curve
y = 1778.6x + 34596
R2 = 0.9945
0
500000
1000000
1500000
2000000
0 200 400 600 800 1000 1200
pg /ml PAR
Net
Mea
n R
LU
Assay Validation Standard Curve: Typical PAR standard curve with trend line. The linear dynamic range is from 10 to 1000 pg/ml.The R2 value for the linear curve fit (shown) was 0.9945.
Net M
ean
RLU
PAR
(pg/
ml)
PAR
(pg/
ml)
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 17
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YPA
RG A
SSAY
14
Poly(ADP-ribose) glycohydrolase (PARG) degrades polymers synthesized by poly(ADP-ribose) polymerase (PARP1). When activated by DNA strand breaks, PARP1 usesNAD+ as a substrate to form ADP-ribose polymers on itself and on specific acceptor proteins.These polymers are in turn rapidly degraded by PARG, a ubiquitously expressed exo- and endoglycohydrolase. PARG may maintain the active state of PARP1 by continuouslyremoving inhibitory ADP-ribose residues from PARP1. The regulation of PARG activity maytherefore influence PARP-mediated cell death. PARG inhibitors could thereby indirectlyinhibit PARP1 activity. Trevigen’s HT PARG Assay Kits measure the loss of biotinylated PARfrom histones attached to strip wells in a 96 well format and is ideal for the screening ofPARG inhibitors and for measuring the activity of PARG in cell extracts.
F E AT U R E S :• Colorimetric or Chemiluminescent readout• 96 well format• Highly sensitive – detects 50 pg PARG per well • Assay Time ~2 hrs
A P P L I C AT I O N S :• Measure PARG Activity in cell extracts• Identify inhibitors and activators of PARG activity• Determination of IC50 values for PARG Inhibitors
K I T C O M P O N E N T S :PARP-HSA and PARG Histone-Coated Strip Wells Buffers and Cocktail Activated DNAStrep HRP and Diluent DEA (known PARG inhibitor)Colorimetric or Chemiluminescent Detection Reagents
R E F E R E N C E S :1. Koh DW, Dawson VL, Dawson TM. 2005. The road to survival goes through PARG.Cell Cycle. 4:397-399.2. Cuzzocrea S, Wang ZQ. 2005. Role of poly(ADP-ribose) glycohydrolase (PARG) in shock,ischemia and reperfusion. Pharmacol Res. 52:100-108.3. Bonicalzi ME, Haince JF, Droit A, Poirier GG. 2005. Regulation of poly(ADP-ribose)metabo-lism by poly(ADP-ribose) glycohydrolase: where and when? Cell Mol Life Sci. 62:739-750.4. Patel NS, Cortes U, Di Poala R, Mazzon E, Mota-Filipe H, Cuzzocrea S, Wang ZQ,Thiemermann C. 2005. Mice lacking the 110-kD isoform of poly(ADP-ribose) glycohydrolaseare protected against renal ischemia/reperfusion injury. J Am Soc Nephrol. 16:712-719.5. Patel CN, Koh DW, Jacobson MK, Oliveira MA. 2005. Identification of three critical acidicresidues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARGcatalytic domain. Biochem J. 388:493-500.6. Falsig J, Christiansen SH, Feuerhahn S, Burkle A, Oei SL, Keil C, Leist M. 2004.Poly(ADP-ribose) glycohydrolase as a target for neuroprotective intervention: assessment ofcurrently available pharmacological tools. Eur J Pharmacol. 497:7-16.7. Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, JacobsonEL, Jacobson MK, Wang ZQ. 2004. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stressin mice. Mol Cell Biol. 24:7163-7178.8. Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, Miwa M. 2004. Loss ofpoly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophilamelanogaster. Proc Natl Acad Sci U S A. 101:82-86.9. Lu XC, Massuda E, Lin Q, Li W, Li JH, Zhang J. 2003. Post-treatment with a novel PARGinhibitor reduces infarct in cerebral ischemia in the rat. Brain Res. 18:99-103.10. Ying W, and Swanson RA. 2000. The poly(ADP-ribose) glycohydrolase inhibitorgallotannin blocks oxidative astrocyte death. Neuroreport. 11:1385-1388.
PARP/ PAR/ PARGHT Universal PARG Assay Kits
4683-096-K $485 HT Universal Colorimetric PARG AssayKit, 96 tests
4682-096-K $485 HT Universal Chemiluminescent PARGAssay Kit, 96 tests
O R D E R I N G I N F O R M A T I O N
ASSAY DESIGN:
Step 1: Ribosylation of histones by PARP.
• Biotinylated-PAR polymer synthesized ontoimmobilized histones using PARP-HSA.
Step 2: PARG degrades PAR. • Biotinylated-PAR attached to histones is hydrolyzed
by PARG.Step 3: Detection to measure remaining biotinylated PARattached to histones via Strep-HRP.• PARG inhibitors are identified by an increase in Signal.
Colorimetric orChemiluminescent
Detection
HRP
Streptavidin
Biotin
PAR
Histones
Trevigen’s Universal PARG Assay
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 18
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
PARP
15
ADP-Ribose Polymerase
Trevigen offers two grades of poly (ADP-ribose) polymerase: “standard” PARP, and highspecific activity (HSA) enzyme. The specific activity serves as a measure of enzyme purity,with the standard enzyme being purified to approximately 50% purity, and the HSA grade togreater than 95% purity. The standard PARP enzyme is useful as a positive control for bothWestern blot analysis and analysis of protein ribosylation. For more detailed studies or fordrug discovery applications, PARP HSA is recommended. Enzyme preparations with lowerspecific activity are available (catalog #s 4667-50-EB and 4667-250-EB). High specific activity PARP (PARP-HSA) is used in Trevigen’s PARP Assay Kits. PARP is activated by 10XActivated DNA(catalog # 4671-096-06), which can be assayed using Trevigen’s UniversalPARP Activation kits (catalog #s 4677-096-K; 4676-096-K see page 9), or PARP ApoptosisAssay kits (catalog #s 4684-096-K; 4685-096-K see page 10).
S O U R C E :Purified from E. coli containing a recombinant plasmid harboring the human PARP gene.
U N I T D E F I N I T I O N :One unit of PARP incorporates 10 femtomoles of NAD+ onto immobilized histones in 30 minutes.High specific activity PARP (PARP-HSA) is provided at 10 units/µl. For enzyme preparations withlower specific activity, (catalog #s 4667-50-EB and 4667-250-EB), the number of units provided(>5 Units/µl) will vary.
A P P L I C AT I O N S : Identification of inhibitors and activators of PARP activityQuantitation of DNA DamageInvestigation of PARP inactivation during apoptosisWestern Blot Analysis
S T O R A G E B U F F E R :20 mM Tris-Cl (pH 8.0), 200 mM NaCl, 1 mM DTT, 0.1% Triton X-100, 50% glycerol, and 0.1mg/ml BSA.
S T O R A G E :Store at -20°C in a manual defrost freezer. For long-term storage, freeze in working aliquots at-80°C. Avoid freeze-thaw cycles.
R E F E R E N C E S :1. Satoh, M.S. and T. Lindahl. 1994. Role of poly(ADP-ribose) formation in DNA repair.Nature 356:356-358.2. Lautier, D., J. Lagueux, J. Thibodeau, L. Menard, and G.G. Poirier. 1993. Molecular andbiochemical features of poly(ADP-ribose) metabolism. Mol Cell Biochem 122:171-193.3. Curtin NJ. 2005. PARP inhibitors for cancer therapy. Expert Rev Mol Med. 7:1-20.4. Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. 2004. NAD+-Dependent Modulation ofChromatin Structure and Transcription by Nucleosome Binding Properties of PARP-1.Cell 119:803–814.5. Virag, L., and Szabo, C. 2002. The therapeutic potential of Poly(ADP-Ribose) Polymeraseinhibitors. Pharmacological Reviews 54:375-429.6. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, CurtinNJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434:913-917.7. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, DillonKJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. 2005. Targeting theDNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917-921.8. Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H,Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. 2009.Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers.N Engl J Med 361:123-134.
PARP/ PAR/ PARGPARP
4668-100-01 $245PARP Enzyme, High Specific Activity,1,000 units
4668-500-01 $850PARP Enzyme, High Specific Activity,5,000 units
4668-02K-01 $2,900PARP Enzyme, High Specific Activity,20,000 units
4667-50-EB $220Human PARP Enzyme and Buffer, 50 µl
4667-250-EB $750Human PARP Enzyme and Buffer, 250 µl
4671-096-06 $8510X Activated DNA, 300 µl
O R D E R I N G I N F O R M A T I O N
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 19
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YPA
RG
16
PARP/ PAR/ PARGPARG
Poly(ADP-ribose) glycohydrolase
Poly(ADP-ribose) glycohydrolase (PARG) degrades poly(ADP-ribose) (PAR) polymerssynthesized by poly(ADP-ribose) polymerase (PARP-1). PARG is ideal for use as a positivecontrol in Trevigen’s PARG Assay Kit (catalog # 4682-096-K and 4683-096-K see page 14)and for Western blot analysis of PARG in cell extracts.
S O U R C E :Purified from E. coli containing a recombinant plasmid harboring the catalytic domain of thebovine PARG gene.
M E A S U R E M E N T O F A C T I V I T Y:PARG activity is measured by the loss of biotinylated PAR from immobilized histones in 30minutes.
A P P L I C AT I O N S :• Identification of inhibitors and activators of PARG activity• Western Blot Analysis
S T O R A G E B U F F E R :20 mM KPO4 (pH 7.2), 50 mM KCl, 0.1 mg/ml BSA, 0.1% Triton® X-100, 1mM DTT,50% Glycerol.
S T O R A G E :Stable for at least one year when stored at −20 °C.
R E F E R E N C E S :1. Koh DW, Dawson VL, Dawson TM. 2005. The road to survival goes through PARG. Cell Cycle 4:397-399.2. Oei SL, Keil C, Ziegler M. 2005. Poly(ADP-ribosylation) and genomic stability.Biochem Cell Biol 83:263-269.3. Cuzzocrea S, Wang ZQ. 2005. Role of poly(ADP-ribose) glycohydrolase (PARG) in shock,ischemia and reperfusion. Pharmacol Res. 52:100-108.4. Bonicalzi ME, Haince JF, Droit A, Poirier GG. 2005. Regulation of poly(ADP-ribose)metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell Mol Life Sci.62:739-750.5. Patel NS, Cortes U, Di Poala R, Mazzon E, Mota-Filipe H, Cuzzocrea S, Wang ZQ,Thiemermann C. 2005. Mice lacking the 110-kD isoform of poly(ADP-ribose) glycohydrolaseare protected against renal ischemia/reperfusion injury. J Am Soc Nephrol. 16:712-719.6. Patel CN, Koh DW, Jacobson MK, Oliveira MA. 2005. Identification of three critical acidicresidues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARGcatalytic domain. Biochem J. 388:493-500.
4680-096-01 $275 PARG Enzyme (1 µg/ml), 300 µl
O R D E R I N G I N F O R M A T I O N
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 20
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
PAR
17
Poly(ADP-ribose) Polymer
PAR was synthesized using poly(ADP-ribose) polymerase (PARP) in the presence of NAD+,cleaved from PARP, and subsequently purified. The PAR chain length ranges from 2 to 300monomers, and it is recognized by Trevigen’s anti-PAR monoclonal (catalog # 4335-MC-100see page 19) and polyclonal (catalog # 4336-BPC-100 see page 20) antibodies.
A P P L I C AT I O N S :Immunodetection as a positive control in ELISA and Western Blot Analysis.
S T O R A G E :PAR is provided in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA and stored at -80°C.It may be aliquoted to avoid repeated freeze-thawing.
R E F E R E N C E S :1. Shah, G.M., et al. 1995. Methods for biochemical study of poly(ADP-ribose) metabolismin vitro and in vivo. Anal Biochem 227:1-13.2. Affar, E.B., et al. 1998. Immunodot blot method for the detection of poly(ADP-ribose)synthesized in vitro and in vivo. Anal Biochem 259:280-283.3. Menard, L. and G.G. Poirier. 1987. Rapid assay of poly (ADP-ribose) glycohydrolase.Biochem Cell Biol 65:668-673.
PARP/ PAR/ PARGPAR
4336-100-01 $175 Poly (ADP) Ribose (PAR) Polymer,100 µl
O R D E R I N G I N F O R M A T I O N
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 21
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YAN
TI-P
ARP
MON
OCLO
NAL
ANTI
BODY
C2-
10
18
The anti-poly (ADP-ribose) polymerase (PARP) mouse monoclonal antibody may be used forthe analysis of PARP protein levels and the detection of cleavage of PARP in apoptotic cells.During apoptosis, PARP-1 activity initially increases, but later falls due to auto-modificationand cleavage by caspases. The specific rate of change in PARP-1 activity is dependent upona variety of factors including cell type, method of induction of DNA damage or apoptosis, andculture conditions. Specific proteolytic cleavage of PARP has been demonstrated to be areliable marker for apoptosis in a wide variety of cell types, generating 85 kDa and 26 kDafragments.
P H Y S I C A L S TAT E :C2-10 is an IgG1 and is provided in 1X PBS/0.1 mg/ml BSA/0.01% sodium azide.
I M M U N O G E N :Calf Thymus PARP-1
S P E C I F I C I T Y:C2-10 detects the 114 kDa PARP-1 holoenzyme, 85 kDa apoptosis-related, andnecrosis-related 50/ 62/74 kDa cleavage fragments. The antibody cross-reacts with human,monkey, hamster, rat and mouse PARP-1 but not chicken.
A P P L I C AT I O N S :Western Blot Analysis and Immunocytochemistry. For Western blotting, a 1:2000 dilutionusing enhanced chemiluminescence (ECL) or 1:1000 dilution using alkaline phosphatase isrecommended.
C O N T R O L :PARP HSA Enzyme (Catalog # 4668-100-01) available separately.
S T O R A G E :Store at 4°C for at least 1 month, or store aliquots at -20°C.
R E F E R E N C E S :1. Lamarre D, B. Talbot, G de Murcia, C Laplante, Y Leduc, A Mazen, GG Poirier. 1988.Structural and functional analysis of poly(ADP-ribose) polymerase: an immunological study.Biochim. Biophys. Acta 950:147-60.2. Simonin F, JP Briand, S Muller, G de Murcia 1991 Detection of Poly(ADP-ribose)Polymerase in Crude Extracts by Activity-Blot. Anal Biochem. 195:226-31.3. Shah GM, D Poirier, C Duchaine, G Brochu, S Desnoyers, J Lagueux, A Verreault, JCHoflack, JB Kirkland, GG Poirier. 1995 Methods for biochemical study of poly(ADP-ribose)metabolism in vitro and in vivo. Anal Biochem. 227:1-13.
PARP/ PAR/ PARGAnti-PARP Monoclonal Antibody C2-10
Western blot analysis of Wehi, Jurkat and CCRF-CEM celllines untreated (H) and treated (T) with 25 µM Etoposidefor 16 hours at 37oC. Cells were lysed in Tris-Glycine SDSsample buffer at the concentration 1 x 107 cells/ml and10 µl of each lysate were loaded per well of a 4-20% Tris-Glycine gel. Proteins were transferred onto anImmobilon FL membrane and PARP was detected withTrevigen’s anti-PARP (C2-10) antibody followed by anIR800-conjugated secondary antibody. The membrane wasscanned using an Odyssey Infrared Imaging System (Licor).
4338-MC-50 $195 Anti-PARP Monoclonal Antibody(clone C2-10), 50 µg
4668-100-01 $245 PARP Enzyme, High Specific Activity,1000 Units
O R D E R I N G I N F O R M A T I O N
Wehi Jurkat CCRF-CEM
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 22
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
ANTI-PAR MONOCLONAL ANTIBODY
19
C O N T E N T S :Catalog # Description Size4335-MC-100 Anti-PAR monoclonal 100 µl4500-10-P PARP treated protein control 10 µl
This monoclonal antibody is specific for poly(ADP-ribose) (PAR) polymer. It can be used todetect ribosylated proteins by ELISA, Western blot, immunocytochemistry in situ, and for immunopurification. It is supplied with a positive control for ELISA and Western blot analysisconsisting of poly-ADP-ribosylated PARP-1 protein (PARP-PAR).
P H Y S I C A L S TAT E :This antibody is an IgG3a and is provided as purified immunoglobulin from mouse ascites in1X PBS containing 50% glycerol at 1 mg/ml. PARP-PAR is provided at 75 µM in 10 mMTris-HCl (pH 8.0), 1 mM EDTA.
I M M U N O G E N :Purified ADP-ribose polymers between 2 and 50 units long
S P E C I F I C I T Y:The antibody is specific for PAR polymers 2 to 50 units long, but does not recognizestructurally related RNA, DNA, ADP-ribose monomers, NAD+, or other nucleic acid monomers.
A P P L I C AT I O N S :ELISA, Western Blot Analysis, immunoprecipitation, and immunopurification. For Westernblots, an antibody dilution of 1:1000 is recommended.
S T O R A G E :Store the anti-PAR monoclonal at -20°C (manual defrost freezer). Store the PARP-treatedprotein control at -80°C.
R E F E R E N C E S :1. Kupper, J.H., G. de Murcia, and A. Burkle. 1990. Inhibition of poly(ADP-ribosyl)ation byoverexpressing the poly(ADP-ribose) polymerase DNA binding domain in mammalian cells.J Biol Chem 265:18721-18724.2. Malik, N., M. Miwa, T. Sugimura, P. Thraves, and M. Smulson. 1983. Immunoaffinityfractionation of the poly(ADP-ribosyl)ated domains of chromatin. PNAS USA 80:2554-2558.3. Kawamitsu, H., H. Hoshino, H. Okada, M. Miwa, H. Momoi, and T. Sugimura. 1984.Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures.Biochem 23:3771- 3777.4. Affar, E.B., et al. 1998. Immunodot blot method for the detection of poly(ADP-ribose)synthesized in vitro and in vivo. Anal Biochem 259:280-283.5. Ismail, I.H., S. Nystrom, J. Nygren, and O. Hammarsten. 2005. Activation of ataxiatelangiectasia mutated by DNA strand break-inducing agents correlates closely with thenumber of DNA double strand breaks. J Biol Chem 280:4649-4655.6. Gagné, J-P., M. Isabelle, K.S. Lo, S. Bourassa, M.J. Hendzel, V.L. Dawson, T.M. Dawson,and G.G. Poirier. 2008. Proteome-wide identification of poly (ADP-ribose) binding proteinsand poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36:6959-6976.
PARP/ PAR/ PARGAnti-PAR Monoclonal Antibody
Western blot analysis of Wehi and Jurkat cells usingTrevigen’s anti-PAR monoclonal antibody. Cells were lysedin Tris-Glycine SDS sample buffer at the concentration 1x107 cells/ml and 10 µl of each lysate were loaded per wellon 4-20% Tris-Glycine gel. Proteins were transferred ontoan Immobilon FL membrane and ribosylated proteins weredetected with Trevigen’s anti-PAR antibody (catalog # 4335-MC-100) followed by IR800-conjugated secondary antibody(Licor). For an absorption control (right panel), the anti-PARantibody was pre-incubated with PAR polymer (#4336-100-01). Membranes were scanned using an Odyssey InfraredImaging System (Licor).
4335-AMC-050 $365 Anti-PAR Monoclonal Affinity Purified,50 µl
4335-MC-100 $260 Anti-PAR Monoclonal Antibody, 100 µl
4335-MC-100-AC $260 Anti-PAR Monoclonal Antibody, 100 µl,and PARP-PAR control protein, 10 µl
4335-MC-01K-AC $2,205 Anti-PAR Monoclonal Antibody, 1000 µl,and PARP-PAR control protein, 100 µl
4500-10-P $75 PARP treated protein control, 10 µl
Western blot analysis of parylated PARP (catalog # 4500-10-P). 15 µl of a 1:10 dilution of PARP-PAR in sample bufferwere separated on a 4-20% Tris-Glycine SDS-PAGE gel andblotted on to PVDF membrane.
Ribosylated proteins were detected with 1:1000 dilutionof anti-PAR followed by a 1:5000 dilution of HRPconjugated goat anti-mouse IgG and visualized usingchemiluminescence.
200>
O R D E R I N G I N F O R M A T I O N
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 23
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YAN
TI-P
AR R
ABBI
T PO
LYCL
ONAL
ANT
IBOD
Y
20
4336-APC-050 $365Anti-PAR Polyclonal Affinity Purified, 50 µl
4336-BPC-100 $315Anti-PAR Polyclonal Antibody, 100 µl
O R D E R I N G I N F O R M A T I O N
A rabbit polyclonal antibody raised against poly(ADP-ribose) (PAR) polymer. The anti-PARpolyclonal antibody can be used to detect ribosylated proteins by immunodetection.Trevigen’s PARP treated control protein (4500-10-P see page 19) and PAR polymer(catalog # 4336-100-01 see page 17) may be used as positive controls.
P H Y S I C A L S TAT E :This polyclonal antibody is a purified IgG fraction in 1X PBS containing 50% glycerol.
I M M U N O G E N :Poly(ADP-ribose) polymer
S P E C I F I C I T Y:This polyclonal antibody detects free PAR and poly-ribosylated proteins.
A P P L I C AT I O N S :For Western and dot blotting, an antibody dilution of 1:1000 is recommended. For ELISA a1:4000 antibody dilution is recommended. Empirical determination of antibody dilutions willbe required for optimum results.
S T O R A G E :-20°C (manual defrost freezer)
R E F E R E N C E S :1. Affar, E.B., et al. 1998. Immunodot blot method for the detection of poly (ADP-ribose)synthesized in vitro and in vivo. Anal. Biochem. 259:280-283. 2. Shah, G.M., et al. 1995. Methods for biochemical study of poly (ADP-ribose) metabolismin vitro and in vivo. Anal. Biochem. 227:1-13.3. Kinders JK, Hollingshead M, Khin S, Rubinstein L, Tomaszewski JE, Doroshow JH, Parchment RE, and the National Cancer Institute Phase 0 Clinical Trials Team. 2008. Preclinical Modeling of a Phase 0 Clinical Trial: Qualification of a Pharmacodynamic Assay ofPoly (ADP-Ribose) Polymerase in Tumor Biopsies of Mouse Xenografts. Clin Cancer Res14:6877-85.
PARP/ PAR/ PARGAnti-PAR Rabbit Polyclonal Antibody
Western blot analysis of Healthy and Etoposide treated Wehicells using Trevigen’s anti-PAR polyclonal antibody.
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 24
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
BIOTIN-NAD+
21
6-Biotin-17-NAD+
Biotinylated NAD+ provides a convenient non-isotopic alternative to radiolabeled NAD+
for determination of IC50 values for candidate PARP inhibitors and studies requiringthis substrate. A number of proteins, including poly (ADP-ribose) polymerase (PARP),use NAD+ as a substrate for their function. The PARP enzyme catalyzes thepolymerization of ADP-ribose onto a number of nuclear proteins. Biotinylated NAD+
allows an indirect measure of PARP activity when biotin incorporation is detected usinga conjugated-streptavidin detection system.
C O N C E N T R AT I O N :250 µM in waterM O L E C U L A R W E I G H T:1180.08 g/mole (1158.09 g/mole as a free acid)
E X T I N C T I O N C O E F F I C I E N T:22,000 at 265 nm
P U R I T Y:Greater than 95% by HPLC
S T O R A G E :Avoid repeated freeze thaw cycles, prepare aliquots and store at -80°C.
R E F E R E N C E S :1. Zhang J, Snyder SH. 1993. Purification of a nitric oxide-stimulated ADP-ribosylatedprotein using biotinylated beta-nicotinamide adenine dinucleotide. Biochemistry 32:2228-33.2. http://www.trevigen.com/MarketingFlyers/JDS_PARP2008_Poster.pdf
PARP/ PAR/ PARGBiotin-NAD+
S
NHHN
HN
OO
NH
N
N
N
NO
OH OH
P
O
O-OP
O
O-O O
OH
Na+
HOO
N+
ONH
2
NHo
HN
O
4670-500-01 $295 Biotin-NAD+, 500 µl
O R D E R I N G I N F O R M A T I O N
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 25
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YFL
UORE
SCEI
N-NA
D+
22
Fluorescein-NAD+ (6-fluorescein-17-nicotinamide-adenine-dinucleotide) Fluorescein-NAD+ provides a convenient, non-isotopic alternative to radiolabeled NAD foruse with enzymes requiring NAD+ as a substrate or cofactor. A number of proteins,including poly (ADP-ribose) polymerases (e.g. PARP-1, PARP-2), and the SIR2 family ofNAD+-dependent histone/protein deacetylases, use NAD+ as a substrate for their function.Fluorescein-conjugated NAD+ permits the direct measurement of PARP and otherNAD+-dependent enzymes by fluorescence microscopy. Fluorescein-NAD+ enters cells withthe aid of Trevigen’s Cell Permeabilization Solution (catalog # 4674-250-01).
P H Y S I C A L S TAT E :Provided in solution at a concentration of 250 µM in deionized water. There is a1:1 stoichiometry for incorporation; one Fluorescein label for each NAD+ molecule.
E X T I N C T I O N C O E F F I C I E N T:38,000 at 262 nm
S T O R A G E :Store at - 80°C
A P P L I C AT I O N S :• Activity measurements of NAD+-requiring enzymes.• Assays to identify inhibitors of activators of NAD+-requiring enzymes. *Cell Permeabilization Solution (catalog # 4674-250-01), is required for in-cell assays for Fluorescein-NAD+ entry.
R E F E R E N C E S :1. Bakondi E, Bai P, Szabó E E, Hunyadi J, Gergely P, Szabó C, Virág L. (2002) Detection ofpoly(ADP-ribose) polymerase activation in oxidatively stressed cells and tissues usingbiotinylated NAD+ substrate. J Histochem Cytochem 50:91-8.
Cell Permeabilization Solution
Cell Permeabilization Solution allows for the efficient transfer of Fluorescein-conjugatedNAD+ across cellular membranes. The solution contains a selective cell permeable peptideto deliver Trevigen’s Fluorescein-NAD+ (catalog # 4673-500-01) from the outside to theinside of intact cells.
P H Y S I C A L S TAT E :Peptide provided as solution in deionized water.
S T O R A G E :Store at -20°C
A P P L I C AT I O N S :• In-cell activity measurements of NAD+-requiring enzymes.• In-cell Assays to identify inhibitors of activators of NAD+-requiring enzymes.
R E F E R E N C E S :1. Fischer, R., Fotin-Mleczek, M., Hufnagel, H. and Brock, R. (2005), Break on through to theOther Side—Biophysics and Cell Biology Shed Light on Cell-Penetrating Peptides.ChemBioChem, 6: 2126–2142.
PARP/ PAR/ PARGFluorescein-NAD+
4673-500-01 $325 Fluorescein-NAD+, 250 µl
4674-250-01 $225 Cell Permeabilization Solution, 250 µl
O R D E R I N G I N F O R M A T I O N
Light
Fluorescein
Merge
H202 3-AB
Wehi cells treated with hydrogen peroxide or 3-AB (PARP inhibitor)were incubated in a PARP cocktail containing Fluorescein-NAD+ andCell Permeabilization Solution were visualized by light and fluorescentmicroscopy.
Fluorescent Image of In-Cell Assay
Incorporation of Fluorescein-labeled ribose from Fluorescein-NAD+ by Poly (ADP-ribose) polymerase (PARP)in presence of Evan’s Blue.
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 26
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
PARRP AND PARG INHIBITORS
23
3-AminobenzamideCatalog # 4667-50-033-Aminobenzamide is a poly(ADP-ribose) polymerase (PARP) inhibitor in the family ofbenzamides. 3-aminobenzamide attenuates tissue injury following transient ischemia andacts as a neuroprotectant since it inhibits PARP, an enzyme activated by peroxynitriteactivity. This PARP inhibitor, with low in vivo toxicity, has been analyzed in the range of 0.1to 10,000 µM, and is provided at 200 mM in ethanol for subsequent serial dilution and usewith Trevigen’s HT Universal PARP Assay Kits.
4-Amino-1,8-naphthalimideCatalog #4667-50-094-amino-1,8-naphthalimide is a potent inhibitor of poly (ADP-ribose) polymerase (PARP)and also reduces ischemia-reperfusion injury in the heart and skeletal muscle. It is1000-fold more potent that 3-aminobenzamide and exhibits mixed-type inhibition withrespect to the substrate NAD+, at micromolar concentrations. This inhibitor, which hasbeen analyzed at a concentration of 20 µM, is provided at 800 µM in DMSO for subsequentserial dilution and use with Trevigen’s HT Universal PARP Assay Kits.
6(5H)-PhenanthridinoneCatalog #4667-50-106(5H)-Phenanthridinone strongly inhibits poly (ADP-ribose) polymerase (PARP) anddisplays immunosuppressive activity. It is a mixed-type inhibitor that acts on both theenzyme and enzyme-NAD+ complex at site(s) distinct from the NAD+-binding site toattenuate decreases in NAD+ and ATP and, consequently, improve cell survival. It alsoinhibits concanavalin A-induced lymphocyte proliferation at micromolar concentrations.This inhibitor, which has been analyzed in the range of 0.18-0.39 µM, is provided at 160µM in DMSO for use with Trevigen’s HT Universal PARP Assay Kits.
BenzamideCatalog #4667-50-11Benzamide is the most potent poly (ADP-ribose) polymerase (PARP) inhibitor in the familyof benzamides. It also acts as a neuroprotectant by inhibition of PARP. Benzamide is twiceas active as its commonly used counterpart 3-aminobenzamide, in delaying or suppressingPARP activation. It is able to prevent nuclear fragmentation and apoptotic-body formationwithout affecting DNA fragmentation during apoptosis. This PARP inhibitor, which hasbeen analyzed in the range of 100 to 500 µM, is provided at 8 mM in ethanol for use withTrevigen’s PARP Assay Kits.
DEACatalog #4680-096-03DEA (6,9-diamino-2-ethoxyacridine lactate monohydrate) is provided at 100 mM in DMSOas a control PARG inhibitor. DEA will inhibit the activity of PARG at a wide range ofconcentrations from 1 µM to 1 mM. This inhibitor is a component of the HT UniversalPARG Assay Kits.
PARP/ PAR/ PARGPARP and PARG Inhibitors
4667-50-03 $75 3-Aminobenzamide, 60 µl
4667-50-09 $110 4-Amino-1,8-naphthalimide, 100 µl
4667-50-10 $110 6(5H)-Phenanthridinone, 100 µl
4667-50-11 $110 Benzamide, 100 µl
4680-096-03 $40DEA, 200 µl
O R D E R I N G I N F O R M A T I O N
Graphical representation of the chemiluminescent readout of the PARPstandard curve and inhibition curve for 3-aminobenzamide using HTUniversal Chemiluminescent PARP Assay Kit (4676-096-K).Each pointrepresents the mean value fro triplicate determinations.
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 27
-
1-800-873-8443 • www.trevigen.com
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YRE
AGEN
T KI
TS
24
Results of a typical CometAssay® experiment in alkalielectrophoresis conditions. DNA fragmentation associatedwith oxidative DNA damage was visualized using theCometAssay® kit. (A) Damaged cell showing characteristicmigration of DNA. (B) Untreated cell showing no DNA damage.
A
B
CometAssay®CometAssay® Reagent Kits for Single CellGel Electrophoresis AssayTrevigen’s CometAssay®, or single cell gel electrophoresis assay, provides a simple andeffective method for evaluating DNA damage in cells. The principle of the assay is basedupon the ability of denatured, cleaved DNA fragments to migrate out of the nucleoid underthe influence of an electric field, whereas undamaged DNA migrates slower and remainswithin the confines of the nucleoid when a current is applied. Evaluation of the DNA “comet”tail shape and migration pattern allows for assessment of DNA damage. The NeutralCometAssay® is typically used to detect double-stranded breaks, whereas the AlkalineCometAssay® is more sensitive, and is used to detect smaller amounts of damage includingsingle and double-stranded breaks.
A S S AY D E S I G N F O R A L K A L I N E C O M E T:Step 1: Cells mixed with low melting agarose are immobilized on specially treated slidesand lysed. • Membranes and histones removed from the DNA of individually isolated cells.Step 2: Samples are treated with alkali to unwind and denature the DNA. • For detection of single and double-stranded breaks.Step 3: Following alkaline electrophoresis slides are stained and visualized byfluorescent microscopy. • Increase in DNA damage results in smaller fragments migrating faster in an electric field. • Quantitative measurements of DNA damage
A P P L I C AT I O N S :• Applicable for the analysis of either single strand or double strand DNA breaks• Study cellular responses to DNA damage• Screen for inhibitors of DNA repair• Test chemicals for toxicity• Screen for environmental mutagens• Probe for specific types of DNA damage using DNA repair glycosylases
C O M P O N E N T S :Catalog # Description4250-050-01 Lysis Solution4250-050-02 Comet LMAgarose4250-050-03 CometSlide™ (2 well)4250-050-04 200 mM EDTA4250-050-05 SYBR® Green
R E L AT E D P R O D U C T S :CometAssay® Electrophoresis SystemCometAssay® Control CellsCometAssay® Silver Staining KitCometSlide™ 2, 20 and 96 Well SlidesCometSlide™ Rack SystemFLARE™ Assays and FLARE™ Slide (3 well)
S T O R A G E :Store components at -20˚C, 4˚C, and room temperature.
4250-050-01 $70CometAssay® Lysis Solution,2x500 ml
4250-050-02 $40CometAssay® LMAgarose,15 ml
4250-050-05 $25SYBR® Green, 5 µl
4250-050-K $225 CometAssay® Kit, 50 Samples(twenty-five 2 well slides)
4252-040-K $225CometAssay® Kit, 40 Samples(two 20 well slides)
4253-096-K $225CometAssay® Kit, 96 Samples(one 96 well slide)
O R D E R I N G I N F O R M A T I O N
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:18 PM Page 28
-
D N A D A M A G E A N D G E N O M I C I N S TA B I L I T YO
VE
RV
IEW
00
1-800-873-8443 • www.trevigen.com
REAGENT KITS
25
CometAssay®CometAssay® Reagent Kit for Single CellGel Electrophoresis AssayR E F E R E N C E S :1. Lemay, M. and K.A. Wood. 1999. Detection of DNA damage and identification ofUV-induced photoproducts using the CometAssay™ kit. BioTechniques 27:846-851.2. Singh, N.P., R.E. Stephens, and E.L. Schneider. 1994. Modifications of alkaline microgelelectrophoresis for sensitive detection of DNA damage. Int J Radiat Biol 66:23-28.3. Collins, A.R., S.J. Duthie, and V.L. Dobson. 1993. Direct enzymatic detection ofendogenous oxidative base damage in human lymphocyte DNA.Carcinogenesis 14:1733-1735.4. Collins, A.R. 2004. The comet assay for DNA damage and repair: principles,applications, and limitations. Mol Biotechnol 26:249-261.5. Olive, P.L. 2002. The comet assay. An overview of techniques.Methods Mol Biol 203:179-194.6. Horvathova, E., M. Dusinska, S. Shaposhnikov, and A.R. Collins. 2004. DNA damageand repair measured in different genomic regions using the comet assay with fluorescentin situ hybridization. Mutagenesis 19:269-276.7. Hartmann, A., M. Schumacher, U. Plappert-Belbig, P. Lowe, W. Suter, and L. Mueller.2004. Use of the alkaline in vivo comet assay for mechanistic genotoxicity investigations.Mutagenesis 19:51-59.
S Y B R ® G R E E N I N U C L E I C A C I D G E L S TA I N L I C E N S I N G T E R M S :This product is sold under license from Molecular Probes, Inc. under US Patents Nos. 5,436,134and 5,658,751 for use in a comet assay for internal research and development only, whereresearch and development use expressly excludes the use of this product for providing medical,diagnostic or any other testing analysis or screening services or providing clinical informationor clinical analysis, in return for compensation on a per-test basis, and research and developmentuse expressly excludes incorporation of this product into another product for commercializationeven if such other product would be commercialized for research and/or development use.
1. Cells mixed with low melting agarose at 37oC (LM Agarose)2. Immobilize cells on CometSlide™3. Treat cells with Lysis Solution (removes membranes and histones from the DNA)4. Samples treated with alkali (unwinds and denatures DNA)5. Samples stained with intercalating dye and visualized by epifluorescence microscopy following alkaline electrophoresis, which reveals DNA breaks
USA_Trevigen_Catalog_2011_1-24:Layout 1 1/24/11 5:19 PM Page 29




![Cell Phones[1] 1](https://static.fdocuments.us/doc/165x107/577d21d11a28ab4e1e95f42a/cell-phones1-1.jpg)