University of Groningen The Staphylococcus aureus secretome … · 2016. 3. 9. · toxic shock and...
Transcript of University of Groningen The Staphylococcus aureus secretome … · 2016. 3. 9. · toxic shock and...

University of Groningen
The Staphylococcus aureus secretomeSibbald, M.J.J.B.
IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite fromit. Please check the document version below.
Document VersionPublisher's PDF, also known as Version of record
Publication date:2010
Link to publication in University of Groningen/UMCG research database
Citation for published version (APA):Sibbald, M. J. J. B. (2010). The Staphylococcus aureus secretome. s.n.
CopyrightOther than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of theauthor(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons).
Take-down policyIf you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediatelyand investigate your claim.
Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons thenumber of authors shown on this cover page is limited to 10 maximum.
Download date: 25-04-2021

53
Chapter 3
Proteogenomics uncovers extreme heterogeneity in the
Staphylococcus aureus exoproteome due to genomic plasticity
and variant gene regulation
A.K. Ziebandt, M. Degner, M.J.J.B. Sibbald, J.P. Arends, M.A. Chlebowicz,
H. Kusch, D. Albrecht, R. Pantuček, J. Doškar, W. Ziebuhr, B.M. Bröker, M. Hecker,
J.M. van Dijl, and S. Engelmann
Submitted for publication, in revision

Chapter 3
54
Summary
Sequencing of at least thirteen S. aureus isolates has shown that genomic plasticity
impacts significantly on the repertoire of virulence factors. However, genome
sequencing does not reveal which genes are de facto expressed by individual isolates.
Here, we have therefore performed a first comprehensive survey of the composition and
variability of the S. aureus exoproteome following a proteogenomics approach. This
involved multi locus sequence typing, virulence gene and prophage profiling by
multiplex PCR, and proteomic analyses of secreted proteins using two-dimensional
protein gel electrophoresis. Dissection of the exoproteomes of 25 clinical isolates revealed
that only seven out of 63 identified secreted proteins were produced by all isolates,
indicating a remarkably high exoproteome heterogeneity within one bacterial species.
The observed variations were caused by both genome plasticity and an unprecedented
variation in gene expression. Our data imply that genomic studies focussing on virulence
gene conservation patterns need to be complemented by protein expression analyses to
assess the full virulence potential of bacterial pathogens like S. aureus. Importantly, the
extensive variability of secreted virulence factors in S. aureus also suggests that the
development of protective vaccines against this pathogen requires a carefully selected
combination of invariably expressed antigens.

Proteogenomics uncovers extreme heterogeneity in the Staphylococcus aureus
exoproteome due to genomic plasticity and variant gene regulation
55
Introduction
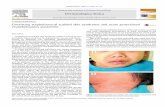
Staphylococcus aureus causes a wide variety of human infections ranging from superficial lesions to severe systemic diseases. Up to one third of the human population carries S. aureus as a commensal bacterium without developing any clinical symptoms. Nevertheless, the colonizing strain can serve as an endogenous reservoir for infection, the incidence of bacteremia being less than 0.02% (von Eiff et al., 2001). Individuals colonized with S. aureus have thus a higher risk to develop an S. aureus infection, but they are also more likely to overcome the disease (Wertheim et al., 2004). It is undisputed that the incidence and outcome of invasive staphylococcal diseases strongly depend on host factors, in particular the immune status of a patient. Yet, the species S. aureus is highly diverse and up to 30% of the genomes of different isolates consist of variable regions, such as pathogenicity islands, lysogenic bacteriophages, and plasmids (Witney et al., 2005). Since these genetic elements encode the majority of virulence factors, it is generally thought that their presence critically determines the clinical symptoms and outcome of an S. aureus infection. However, virulence-associated genes may also be located in the core genome as illustrated by the spa, aur, hla, lip, clfAB,
map/eap, fnbA, and coa genes (Holden et al., 2004; Peacock et al., 2002). It is generally accepted that certain strains are more virulent than others (Melles et al., 2004), although, under certain conditions, any S. aureus genotype may have the potential to induce life-threatening infections. This suggests that there is no simple relationship between bacterial genotype and clinical outcome. Apart from several well-defined toxin-mediated diseases (e.g. toxic shock and scalded skin syndrome, necrotizing pneumonia) (Musser et al., 1990; Gemmell, 1995; Labandeira-Rey et al., 2007), it remains difficult to predict the onset and course of an S. aureus infection in a given patient solely on the basis of the virulence gene repertoire of the strain involved. In fact, the vast majority of severe S. aureus infections are probably caused by the concerted action of multiple virulence factors. Molecular typing and genome analyses of clinical isolates focussing on virulence gene distribution have been major steps towards a thorough understanding of these complex phenomena. Also, there is growing evidence that the presence and activities of certain regulators play a role in the variation of virulence gene expression in clinical isolates (Blevins et al., 2002; Karlsson and Arvidson, 2002). Here, we complement the genetic identification of virulence factors in different S. aureus isolates with proteome analyses of secreted proteins, which represent an important reservoir of virulence factors. More specifically, in this proteogenomics approach we have addressed the questions (i) of whether particular virulence genes are expressed in different isolates and, if so, (ii) in what quantities? The results reveal an unprecedented heterogeneity in the analyzed S. aureus exoproteomes, which is caused by both genomic plasticity and an amazing variability in the levels of gene expression.
Materials and Methods
Bacterial strains
Twenty five S. aureus isolates derived from a variety of human infections were collected by the university hospital in Groningen. All strains were identified as S. aureus by plating and coagulase tests. Resistance against antibiotics was determined by routine disk diffusion assays. For RNAIII

Chapter 3
56
transcription analyses, S. aureus RN6390, COL, and Newman were used as reference strains (Shafer and Iandolo, 1979; Duthie and Lorenz, 1952; Novick, 1967).
Multilocus sequence typing (MLST) MLST was performed according to the protocol described by Enright et al. (Enright et al., 2000). The obtained sequences for each locus were submitted to the Internet database (www.mlst.net) and the resulting allelic profiles were assigned to particular sequence types (ST) for each isolate. The eBURST (Based upon related sequences) algorithm software was used to classify different sequence types into clusters of clonal complexes (CC). Detection of prophages and virulence genes using multiplex PCR
Genomic DNA was extracted from S. aureus clinical isolates and used as a template in multiplex PCR assays targeting structural prophage genes of head-tail modules described previously (Pantucek et al., 2004). Three different phage types (A-like, B-like, and F-like) corresponding to putative phage species 3A, 11, and 77, respectively, were thus identified. The F-like phages include clearly distinguishable subgroups Fa and Fb, while the B-like phages include 5 subgroups Ba – Be of different phages, in which the packaging, head, and tail genes belong to different modules. Therefore, three PCR assays were used for phage identification (Table 1).
Table 1. Oligonucleotides used in this study
Target Primer Sequence 5´- 3´ Reference
S. aureus, positive control
SAU1 SAU2
GACGGCTTTGATGGCTAGTGG AGTTAATTCACGCCCTAGTG
(Pantucek et al., 2004)
A-like phage, tail SGA1 SGA2
TATCAGGCGAGAATTAAGGG CTTTGACATGACATCCGCTTGAC
(Pantucek et al., 2004)
B-like phage (all subgroups), tail
SGB1 SGB2
ACTTATCCAGGTGGYGTTATTG TGTATTTAATTTCGCCGTTAGTG
(Pantucek et al., 2004)
F-like phage (both subgroups), tail
SGF1 SGF2
CGATGGACGGCTACACAGA TTGTTCAGAAACTTCCCAACCTG
(Pantucek et al., 2004)
B-like phage, subgroup Ba portal
SGBa1 SGBa2
AAGATGATAACTTTAGTGGCAC TCATTGATGTYTCTAGGGTC
this study
B-like phage, subgroup Bb portal
SGBb1 SGBb2
CTGATTATGTGTACGCAGAG TTCCGTTAAACTCGTCAGA
this study
B-like phage, subgroup Bc portal
SGBc1 SGBc2
TTGTTAAGGAACCYAAGCC GCCTCTAATTCTTCGTGCTC
this study
B-like phage, subgroup Bd portal
SGBd1 SGBd2
AAGTTACGTCGCTGGC GCTTGTTCTGCTGGCACTCT
this study
B-like phage, subgroup Be portal
SGBe1 SGBe2
AAATGAAACTATTCCGTGTT AAYGCTATAAAYGGYACTCT
this study
F-like phage, subgroup Fa portal
SGFa1 SGFa2
TACGGGAAAATATTCGGAAG ATAATCCGCACCTCATTCCT
(Pantucek et al., 2004)
F-like phage, subgroup Fb portal
SGFb1 SGFb2
AGACACATTAAGTCGCACGATAG TCTTCTCTGGCACGGTCTCTT
(Pantucek et al., 2004)
Phage integrase ФSa1 (phage ETA)
phiSa1-F phiSa1-R
AAGCTAAGTTCGGGCACA GTAATGTTTGGGAGCCAT
this study
Phage integrase ФSa2 (phage 12)
phiSa2-F phiSa2-R
TCAAGTAACCCGTCAACTC ATGTCTAAATGTGTGCGTG
this study
Phage integrase ФSa3 (phage 13)
phiSa3-F phiSa3-R
GAAAAACAAACGGTGCTAT TTATTGACTCTACAGGCTGA
this study
Phage integrase ФSa4
phiSa4-F phiSa4-R
ATTGATATTAACGGAACTC TAAACTTATATGCGTGTGT
this study
Phage integrase ФSa5 (phage 11)
phiSa5-F phiSa5-R
AAAGATGCCAAACTAGCTG CTTGTGGTTTTGTTCTGG
this study
Phage integrase ФSa6 (phage L54a)
phiSa6-F phiSa6-R
GCCATCAATTCAAGGATAG TCTGCAGCTGAGGACAAT
this study
Phage integrase ФSa7 (phage 96)
phiSa7-F phiSa7-R
AAACTAAAGCTGAGGCAAC TCATTAGTACGACCTCGAC
this study
Phage integrase ФSa8 (phage 53)
phiSa8-F phiSa8-R
GTCCGGTAGCTAGAGGTC GGCGTATGCTTGACTGTGT
this study

Proteogenomics uncovers extreme heterogeneity in the Staphylococcus aureus
exoproteome due to genomic plasticity and variant gene regulation
57
Target Primer Sequence 5´- 3´ Reference hlb hlb-2
hlb-527 AGCTTCAAACTTAAATGTCA CCGAGTACAGGTGTTTGGTA
(Goerke et al., 2006)
sea Sea-1 Sea-2
AGATCATTCGTGGTATAACG TTAACCGAAGGTTCTGTAGA
(Van Wamel et al., 2006)
sep Sep-1 Sep-2
AATCATAACCAACCGAATCA TCATAATGGAAGTGCTATAA
(Van Wamel et al., 2006)
sak Sak-1 Sak-2
AAGGCGATGACGCGAGTTAT GCGCTTGGATCTAATTCAAC
(Van Wamel et al., 2006)
chp Chip-up Chip-low
CACCATCATTCAGCGAAAG GATATATAAAGGTTTGGCAAG
this study
scn Scin-up Scin-low
AGTCTTTTGACTTAAGAGC GTTTTAGCATCACCACTAGTA
this study
geh gehSA-up gehSA-low
TTCTTAATGGCTACAACGAGT GATAAAATCGACATGATCCC
this study
hlgA hlgA_F hlgA_R
GTCAAGGTGCAGAAATCATC GAAATAGTCTCTTGCTGCTG
this study
splD splD_F splD_R
CCAAAGCAGAAAATAGTGTG CATAACACCAATTGCTTCTC
this study
splE splE_F splE_F
GTGTCGTTTCGATAGGATCT GTTGCCTTTAACTGACAGCA
this study
24636604 24636604_F 24636604_R
GACAGCATCTCACTGTAAAG TCACGCTTTTGACAGATAAG
this study
SAR0422 SAR0422_F SAR0422_R
CAAGTTTAGCATTGGGAATG CGTCGATAGAATCACCCATAC
this study
SAR0435 SAR0435_F SAR0435_R
GTTTAGCACTAGGGATTTTG GGCTCTGTCTTATAAAGTTC
this study
SAR1905 SAR1905_F SAR1905_R
CGATTGCAACTCTTTCATTC CCATACAAAGGCATACTAAG
this study
SA1755 SA1755_F SA1755_R
CTTTTGAACCGTTTCCTACA CAGTATTTTTTGTGCCTTTC
this study
SA0092 SA0092_F SA0092_R
GTAAAGAAGCGGAAGTTAAG GCTTTATTCGTCGGTATATC
this study
SA1001 SA1001_F SA1001_R
CTGCAGGTCTTTTAACTCAA GCTTTCTTCACATCACCTTG
this study
SACOL0478 SACOL0478_F SACOL0478_R
GTTAGATCACAAGCTACTCA GACGTTGATGTAACACTATC
this study
RNAIII RNAIII_F RNAIII_R
AGGAAGGAGTGATTTCAATG CTAATACGACTCACTATAGGGAGAACTCATCCCTTCTTCATTAC
this study
Notably, the nomenclature of the prophage genomes based only on the genes for virion proteins is not absolute, because of their mosaic structure. Therefore, a novel PCR-based molecular assay for identification and classification of S. aureus phage integrase genes was also applied for prophage characterization. For designation of bacteriophage integrase gene families, the updated classification scheme was used which denotes the previously sequenced phages and S. aureus genomes ФSa1 - ФSa5 (Lindsay and Holden, 2004). Since so far unclassified integrases were identified, the new designations ФSa6, ФSa7 and ФSa8 were introduced in this work for integrases of phages L54a (GenBank Accession no. M27965), Ф96 (NC_007057) and Ф53 (NC_007049), respectively. For multiplex PCR, the reaction mixtures (25 µl) consisted of 75 mM Tris-HCl pH 9.0, 50 mM KCl, 2 mM MgCl2, 250 µM
each of dNTPs, primer pairs phiSa1 and phiSa2 (0.25 µM each), phiSa3 (0.2 µM each), phiSa4 and phiSa5 (0.2 µM each), phiSa6, phiSa7 and phiSa8 (0.1 µM each). Primer sequences are listed in Table
1. To each reaction, 1.5 unit Taq DNA polymerase (Invitrogen) and template DNA (50 ng) in a 3 µl volume were added. PCR was performed using 30 cycles of amplification consisting of denaturation (1 min, 94°C), annealing (1 min 30 s, 56°C) and DNA chain extension (1 min, 72°C). Phage associated virulence genes sea, sep, sak, chp, and scn as well as geh and hlb were detected using standard PCR with the primers listed in Table 1. For the analysis of genes encoding superantigens and other extracellular proteins as well as for agr typing, primer pairs were used as shown in Table 1 or described previously (Holtfreter et al., 2004;

Chapter 3
58
Lina et al., 2003). The amplifications were performed with Taq polymerase in a thermocycler with the following conditions: initial denaturation at 95°C for 5 min, followed by 30 stringent cycles (1 min of denaturation at 95°C, 1 min of annealing at the temperature indicated in Table 1, and 1 min of extension at 72°C), and a final extension step at 72°C for 5 min. The quality of the DNA extracts and the absence of PCR inhibitors were confirmed by amplification of glyceraldehyde-3-phosphate dehydrogenase or 16S rRNA. The PCR products were then analyzed by electrophoresis through a 1% agarose gel. At least two independent experiments were performed for each determination.
Extracellular proteins
For the preparation of extracellular protein extracts, bacteria were grown in Tryptic Soy Broth (TSB). At optical densities (OD540) of 10, the extracellular proteins from 100 ml supernatant were precipitated, washed, dried, and resolved as described previously (Ziebandt et al., 2004). The protein concentration was determined using Roti®-Nanoquant according to manufacturer´s instructions (Carl Roth GmbH & Co, Karlsruhe, Germany). Analytical and preparative 2D polyacrylamide gel electrophoresis (PAGE)
Protein extracts (350 µg) were loaded onto commercially available IPG strips (pH 3-10, GE-Healthcare, Uppsala, Sweden). 2D PAGE was performed as described previously (Bernhardt et al., 1999; Eymann et al., 2004). The resulting protein gels were stained with colloidal Coomassie Blue G-250 (Candiano et al., 2004) and scanned with the light scanner. Protein identification
For identification of proteins by Matrix-assisted Laser Desorption Ionization-Time of Flight mass spectrometry (MALDI-TOF MS), Coomassie stained protein spots were excised from gels using a spot cutter (Proteome WorkTM) with a picker head of 2 mm and transferred into 96-well microtiter plates. Digestion with trypsin and subsequent spotting of peptide solutions onto the MALDI targets were performed automatically in an Ettan Spot Handling Workstation (GE-Healthcare, Little Chalfont, United Kingdom) using a modified standard protocol (Eymann et al., 2004). MALDI-TOF MS analyses of spotted peptide solutions were carried out on a Proteome-Analyzer 4700/4800 (Applied Biosystems, Foster City, CA as described previously (Eymann et al., 2004). MALDI-TOF-TOF analysis was performed for the three highest peaks of the TOF spectrum as described previously (Eymann et al., 2004; Wolf et al., 2008). Database searches were performed using the GPS explorer software version 3.6 (build 329) with the organism-specific databases. By using the MASCOT search engine version 2.1.0.4. (Matrix Science, London, UK) the combined MS and MS/MS peak lists for each protein spot were searched against a database containing protein sequences derived from the genome sequences of all sequenced S. aureus strains and, moreover, all additional protein sequences of S. aureus that have been found in publically available databases. Search parameters were as described previously (Wolf et al., 2008). RNA-isolation and dot blot analysis
Total RNA from S. aureus was isolated using the acid-phenol method with some modifications (Fuchs et al., 2007). Digoxigenin-labeled RNA probe for RNAIII was prepared by in vitro transcription with T7 RNA polymerase by using a PCR fragment as template. The PCR fragment was generated by using chromosomal DNA of S. aureus N315 and the respective oligonucleotides (Table 1). Dot blot analyses were carried out by using serial dilutions of total RNA prepared from S. aureus isolates and reference strains grown in TSB medium to an optical density (OD540) of 10. The digoxigenin-labeled RNA probe was used for hybridization. The hybridization signals were detected using a Lumi-Imager and analyzed using the software package Lumi-Analyst (Roche Diagnostics, Mannheim, Germany).

Proteogenomics uncovers extreme heterogeneity in the Staphylococcus aureus
exoproteome due to genomic plasticity and variant gene regulation
59
Results and Discussion
Genetic characterization of clinical isolates
A total of 25 clinical isolates, including eight nosocomial methicillin-resistant S. aureus (MRSA) and three community-acquired MRSA (caMRSA) isolates, were used in this study. The isolates were obtained from 19 patients with different clinical symptoms during a 4.5 year period (from February 2001 to September 2005). Strains were isolated from blood, nose, feet, perineum, fingers, throat, pleural fluid, and umbilicus. Five of these isolates were colonizing strains while 12 isolates came from septic patients, five isolates induced wound infections, four isolates arthritis, three isolates pneumonia, two isolates cholangitis, and three individual isolates were involved in abscess formation, panaritium, or endocarditis. On the basis of strain typing data obtained by Pulsed-Field-Gel-Electrophoresis (PFGE) and Multi-Locus-Sequence-Typing (MLST) the isolates were grouped into eleven different sequence types (Supplemental table IVa). Three new sequence types were detected, i.e. ST869, ST870, and ST903. Moreover, the agr types and the prevalence of enterotoxin genes and other clinically relevant genes (e.g. mecA, pvl, etd, eta) were determined for all isolates by using multiplex PCR (Table 2, Supplemental table IVb). The sea gene (72%) as well as genes belonging to the enterotoxin gene cluster (egc) (44%) were found to be the most prevalent enterotoxin genes, while none of the isolates carried tst-1,
sec, see, seh, or sel. Moreover, pvl was identified in four and etd in two isolates, respectively. Typing of agr revealed that six isolates encoded agr1, thirteen agr2, three agr3 and none of the isolates encoded agr4. Additionally, we analyzed the prophage content of these isolates by a multiplex PCR assay. Altogether we identified 11 different prophages. While most of the isolates contained three prophages, no known prophage was detected in two isolates (N, W) (Table 3). The hlb converting phage Sa3 was the most prevalent phage: in 20 isolates we identified either the Fa-type (16 isolates) or the Fb-type of the Sa3 prophage. This prophage is common among human isolates and often encodes immune evasion molecules (SAK, SCIN, and CHIP) as well as enterotoxins (SEA or SEP) and has previously been detected in collections of clinical strains (Goerke et al., 2006). Moreover, we identified the Fa-type phage Sa1 (1 isolate), the A-type phages Sa2 (7 isolates), Sa4 (1 isolate), and Sa6 (6 isolates), as well as the B- type phages Sa1 (2 isolates), Sa5 (1 isolate), Sa6 (3 isolates), Sa8 (9 isolates), and Sa9 (2 isolates). According to our genetic characterization, the 25 isolates represent 17 clonally divergent strains. Five strains were isolated more than once, either in the same patient or in different patients (Supplemental table IVa). Interestingly, isolates C, D, G, H, I, J, and K might have evolved from just one clone (=G228) which was responsible for a hospital outbreak and caused wound infections in three of the patients included in this study. The exoproteomes of clinical isolates are highly heterogeneous
Proteomics is an extremely powerful tool for analyzing virulence gene expression in multiple clinical isolates. Since the extracellular proteome of a pathogenic bacterium represents a key reservoir of virulence factors, the present study focused on this particular subproteome. All 25 isolates were cultivated in TSB medium and extracellular proteins were prepared from the supernatants at an optical density (OD) at 540 nm of 10. Proteins were separated on 2D gels and protein spots were identified by MALDI-TOF MS/MS or N-terminal sequencing

Chapter 3
60
(Supplemental tables IVb). Comparison of extracellular protein patterns revealed an unanticipated degree of exoproteome heterogeneity among the 17 clonally different strains (Figure 1). Table 2. Virulence genes identified by multiplex PCR in different clinical S. aureus isolates
Isolates
Gene A B C D E F G H I J K L M N O P Q R S T U V W X Y
eta
etd + +
hlgA + + + + + + + + + + + + + + + + + + + + + + + + +
pvl + + + +
SA0092 + + + + + + + + + + + + + + + + + + + + + + +
SA1001 + + + + + + + + + + + + + + + + + + + + + + +
SA1755 + + + + + + + + +
SACOL0478 + + + + + +
SAR0422 + + + + + + + + + + + + + + + + + + + + + + + + +
SAR0435 +
SAR1905 + + +
sea + + + + + + + + + + + + + + + +
seb + + + + + + + + +
sec
sed + +
see
seg + + + + + + + + + + +
seh
sei + + + + + + + + + + +
sej + +
sek + + + +
sel
sem + + + + + + + + + + +
sen + + + + + + + + + + +
seo + + + + + + + + + + +
sep + + +
seq + + + +
ser + +
seu +
splD + + + + + + + + + + + + + + + + + + + + + + + + +
splE + + + + + + + + + + +
tst-1
24636604 + +

Proteogenomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene
regulation
61
Table 3. Prophages identified in different clinical S. aureus isolates Phage integrase Classification of prophages according to
genes for virion proteins Innate immune evasion cluster
pattern of F-like phages
# of
phages
Assumed lysogenic pattern
(type of capsida/integrase class)
Strain Sa1 Sa2 Sa3 Sa4 Sa5 Sa6 Sa7 Sa8 A-like
B- like
F- like
Ba Bb Bc Bd Be Fa Fb hlb sea sep sak chp scn
A + + + + + + + + 3 A-phage/Sa2; B-phage/Sa6; Fa-phage/Sa3 B + + + + + + + + + + + 3 A-phage/Sa2; B-phage/Sa6; Fa-phage/Sa3 C + + + + + + + + + + + 3 A-phage/Sa6; B-phage/Sa8; Fa-phage/Sa3 D + + + + + + + + + + + 3 A-phage/Sa6; B-phage/Sa8; Fa-phage/Sa3 E + + + + + + + + + + + 3 A-phage/Sa2; B-phage/Sa8; Fb-phage/Sa3 F + + + + + + + + + + + 3 A-phage/Sa2; B-phage/Sa8; Fb-phage/Sa3 G + + + + + + 2 A-phage/Sa6; B-phage/Sa8 H + + + + + + + + + + + 3 A-phage/Sa6; B-phage/Sa8; Fa-phage/Sa3 I + + + + + + + + + + + 3 A-phage/Sa6; B-phage/Sa8; Fa-phage/Sa3 J + + + + + + + + + + + 3 A-phage/Sa6; B-phage/Sa8; Fa-phage/Sa3 K + + + + + + + + + + + 3 A-phage/Sa6; B-phage/Sa8; Fa-phage/Sa3 L + + + + + + + + 2 A-phage/Sa2;Fb-phage/Sa3 M + + + + + + + 2 A-phage/Sa2;Fb-phage/Sa3 N + + 0 O + + + + + + + 1 Fa-phage/Sa3 P + + + + + + + + + 2 B-phage/Sa1; Fa-phage/Sa3 Q + + + + + + + + + + + + + 4 A-phage/Sa2; B-phage/Sa5; B-phage/Sa6; Fa-
phage/Sa3 R + + + + + 1 B-phage/Sa8 S + + + + + + 1 Fa-phage/Sa3 T + + + + + + 1 Fa-phage/Sa1 U + + + + + + + + + + 3 A-phage/Sa4; B-phage/not identified; Fa-phage/Sa3 V + + + + + + + + + + 2 B-phage/Sa1; Fa-phage/Sa3 W + 0 X + + + + + + 1 Fa-phage/Sa3 Y + + + + + + 1 Fa-phage/Sa3
a According to R. Pantuček et al. (Pantucek et al., 2004)

Chapter 3
62
Figure 1. Characterization of extracellular proteomes of different clinical S. aureus isolates. Cells were grown
in TSB medium to an OD540 of 10. Proteins in culture supernatants were collected by TCA precipitation. 350 µg of
the protein extract of each strain was separated on 2D gels, using commercially available IPG strips (pH 3-10,
GE-Healthcare, Sweden) for the first dimension. Protein spots were detected by staining with colloidal Coomassie
Brillant Blue. For protein identification, individual protein spots were excised from the gel and digested with
trypsin. The resulting peptide solution was analyzed by tandem mass spectrometry on a Proteome Analyzer
4700/4800 (Applied Biosystems, USA). The respective strain is indicated in the upper left corner of each gel.
Altogether 206 distinct proteins were identified (Supplemental table IVb). 107 of these proteins showed signal peptides typical for Sec-translocated proteins. Using PSORTb software (http://www.psort.org/psortb), 63 of the Sec-translocated proteins were predicted to

Proteogenomics uncovers extreme heterogeneity in the Staphylococcus aureus
exoproteome due to genomic plasticity and variant gene regulation
63
Cytosolic; 72
Extracellular; 63
Cell wall; 19
Membrane; 5
Unknown; 47Cytosolic
Extracellular
Cell wall
Membrane
Unknown
be extracellular, 19 were predicted to be cell wall-bound proteins (Figure 2) and the localization of a further 25 proteins is currently unknown (Sibbald et al., 2006). Moreover, we identified 72 cytoplasmic proteins and 5 membrane proteins. The N-terminal sequences (AEANSSMVSKK and GTTPAAA) of two protein spots did not match any of the protein sequences present in the NCBI and other databases, indicating that our knowledge of extracellular proteins produced by S. aureus is not yet complete.
Figure 2. Predicted localization of extracellular
proteins of 25 clinical S. aureus isolates. A total of
206 distinct proteins were identified in the growth
medium fractions of 25 clinical isolates. For the
prediction of protein localization PSORT software
was used.
Interestingly, only seven out of 63 predicted Sec-dependent extracellular proteins were found to be produced by all 17 clonally different strains (i.e. Aly, IsaA, Lip, LytM, Nuc, SA0620, and SA2097). A further nine proteins (i.e. Aur, Geh, GlpQ, Hla, HlgB, SA0570, SA1812, SspA, SspB) were identified in at least 80% of these strains (Supplemental table IVc). Interestingly, four of the invariant proteins (i.e. IsaA, LytM, SA0620, and SA2097) were recently shown to be regulated by the WalK/WalR two-component system which is essential for cell viability and cell wall metabolism (Dubrac et al., 2007). IsaA and LytM share a conserved transglycosylase/muramidase domain, and SA0620 and SA2097 contain a CHAP amidase domain. All these proteins have an N-terminal cell wall-binding domain in common, indicating that they are exported and possibly non-covalently associated to the cell wall. It is worth noting in this context that IsaA-specific antibodies have previously been detected in healthy adults, colonized patients and patients suffering from sepsis, suggesting that IsaA is also expressed in vivo (Lorenz et al., 2000; Clarke et al., 2006). Most strikingly, the amounts of the invariant proteins varied significantly between individual strains (Figure 3). Expression heterogeneity probably triggered by varying SigB and SarA activities has been observed before for extracellular proteases (Karlsson and Arvidson, 2002). Notably, our present data indicate that this phenomenon is by no means restricted to proteases, but applies to secreted virulence factors in general. 31 proteins were found to be unique for one or two strains under the conditions tested. While the functions of some of these proteins, such as Hlb, LukE, and SEB, in S. aureus-associated virulence are well characterized, the functions of other proteins are less clear and, in many cases, remain to be elucidated. Why were those proteins missing from the exoproteome of the remaining strains? There are at least three possible explanations for this phenomenon: the respective genes (i) are absent, (ii) represent pseudo genes or (iii) are not expressed or expressed in very low amounts. The lack of detection of SED, SEK, SEP, SEQ, SER, Etd, 24636604, and SAR0435 correlated with the absence of their coding genes. By contrast, while HlgA, SplD, SAR0422, SA0092, and SA1001 were encoded by at least 80% of the strains, these proteins were synthesized in detectable amounts only in one or two strains (Table 4).

Chapter 3
64
Figure 3. Relative amounts of extracellular proteins detected in at least 80% of investigated S. aureus isolates.
The respective sector on the 2D gel of each isolate is shown for the proteins indicated. The proteins were stained
with colloidal Coomassie Brillant Blue as described for Figure 1.
Table 4. Identification of genes for which gene products were detected in 20% of the
investigated S. aureus isolatesa Gene A B C D E F G H I J K L M N O P Q R S T U V W X Y
sea + + + + + + + + + + + + + + + + seb + + + + + + + + + sed + + sek + + + + sep + + + seq + + + + ser + + etd + + hlgA + + + + + + + + + + + + + + + + + + + + + + + + + splD + + + + + + + + + + + + + + + + + + + + + + + + + splE + + + + + + + + + + + 24636604 + + SAR0422 + + + + + + + + + + + + + + + + + + + + + + + + + SAR0435 + SAR1905 + + + SA1755 + + + + + + + + + SA0092 + + + + + + + + + + + + + + + + + + + + + + + SACOL0478 + + + + + + SA1001 + + + + + + + + + + + + + + + + + + + + + + +
a genes whose gene product was detected on 2D gels are shaded grey for the respective strains

Proteogenomics uncovers extreme heterogeneity in the Staphylococcus aureus
exoproteome due to genomic plasticity and variant gene regulation
65
Variations in the expression levels of virulence genes may relate to differential activities of specific S. aureus gene regulators. Especially RNAIII has previously been identified as a main regulator of virulence gene expression (Novick, 2003), and the loss of RNAIII was shown to affect the extracellular protein pattern of S. aureus dramatically (Ziebandt et al., 2004). To analyse RNAIII levels, we performed dot blot analyses using RNA prepared from cells grown to an OD540 of 10. This revealed that RNAIII was not produced at detectable levels in 11 isolates (C, D, G, H, I, J, K, T, W, X, Y) (Figure 4). With the exception of isolate W this correlates very well with a diminished expression level of late virulence factors (Figure 1), confirming the general importance of RNAIII in virulence gene expression. At the same time this shows that exceptions are possible as was the case for isolate W. Interestingly, these 11 isolates were involved in wound infection, arthritis or cholangitis, respectively (Supplemental table IVa). In mice, agr mutants were shown to have a growth advantage within a mixed population of S. aureus residing in abscesses and wounds (Schwan et al., 2003). Possibly, reduced RNAIII levels and the resulting decreased expression of RNAIII-dependent virulence genes might be correlated to the induction of some of the observed clinical symptoms. In the 14 remaining isolates, RNAIII transcripts have been detected in varying amounts, which may, at least in part, account for the observed differences in virulence factor production. Similar exoprotein patterns in closely related isolates
While virulence gene expression of clonally different isolates was highly variable, we observed very similar protein expression patterns in closely related isolates (i.e. B398, E8, L80, G228, and X8) (Supplemental table IVa, Figure 1). Isolates L and M (=L80), which belong to the clonal group ST80, displayed all the genetic characteristics (pvl, hlgA, etd,
edinB) as well as characteristic resistance patterns typically found in caMRSA strains with epidemic spread in the European population (Supplemental table IVa, Tables 2 and 3) (Monecke et al., 2006; Vandenesch et al., 2003). The isolates were derived from two patients suffering from panaritium and abscesses, respectively. Interestingly, the extracellular protein pattern of isolates L and M was identical, suggesting that the virulence gene expression pattern may be very stable over extended periods of time (April 2002 - April 2004) (Figure 1). The protein expression profiles of three other isolate pairs, A/B (=B398), E/F (=E8), and X/Y (=X8) were also very similar. The strain pair A/B was identified by MLST as sequence type 398. This clonal lineage of S. aureus is frequently found on pigs in the Netherlands and was recently described to spread from animals to humans (Armand-Lefevre et al., 2005; van Belkum A. et al., 2008; Witte et al., 2007). In the present study the isolates came from a mother and her daughter and were involved in pneumonia and phlegmone, respectively. They encode pvl but none of the known superantigens. As described for other isolates of this lineage, strain typing by PFGE failed, which was possibly due to the activity of a restriction/methylation system typical for this lineage (Bens et al., 2006). Notably, the two isolates of strains B398 differed with respect to mecA gene conservation (Supplemental table IVa), suggesting that the acquisition or loss of methicillin resistance had no significant influence on virulence gene expression (Figure 1). This difference in mecA conservation was also observed for the two X8 strains. As indicated above, one strain (G228), which was responsible for a hospital outbreak, was identified seven times in four different patients. The first isolate (G) was obtained at the end of 2003 and the most recent isolate (I) was identified early in 2005 as the colonizing S. aureus

Chapter 3
66
strain of a patient who suffered from a wound infection induced by this strain (isolate H) one year before. We found the hlb converting phage FaSa3 in all these isolates, except for isolate G, as might be expected from the phage dynamics during infection (Goerke et al., 2006; Goerke et al., 2004). In accordance with this, the phenotype of strain G288 changed from Hlb positive (G) to Hlb negative (C, D, H, I, J, K) (Table 3). However, the prophage did not significantly alter the expression of other virulence-associated genes (Figure 1). This might be mainly due to the fact that RNAIII was not expressed in these isolates under our experimental conditions (Figure 4) and, accordingly, only a few virulence factors were produced.
Figure 4. Transcription of RNAIII in different clinical S. aureus isolates. RNA was prepared from cells grown in
TSB to an optical density of 10. Serial dilutions of total RNA of clinical isolates and reference strains (RN6390,
COL, Newman) were blotted and crosslinked onto the same positively charged nylon membrane. The membrane-
bound RNA was hybridized with a digoxigenin labelled RNA probe complementary to RNAIII. Chemiluminescence
signals were detected with a LumiImager (Roche Diagnostics, Mannheim, Germany).
Concluding remarks
In conclusion, our data show that in the species S. aureus, genome plasticity is only one of several factors involved in exoproteome profile heterogeneity. Expression regulation as well as protein secretion and modification processes add further dimensions to the heterogeneity of the virulence potential of S. aureus. Such effects might be further enhanced and fine-tuned by promoter mutations, differential activities of regulatory molecules and translation regulation mechanisms. Most probably, the profoundly heterogeneous expression pattern of virulence genes observed under identical in vitro conditions reflects a very high degree of variability in
vivo. It seems reasonable to suggest that these different virulence protein patterns are linked to different clinical symptoms in the host. We are currently comparing virulence gene expression profiles of S. aureus isolates that induced very similar symptoms. In this way, we hope to identify symptom-/disease-related proteomic signatures that may help in elucidating specific pathogenic mechanisms. It has been shown that S. aureus carriers mount a very selective and protective antibody response against superantigens of their colonizing strains (Holtfreter et al., 2006). Similarly, specific adaptive immune responses might also be raised against other virulence factors, which vary between strains to a similar extent. These and other immune mechanisms might explain why S. aureus carriers with bacteremia generally have a better outcome than non-carriers (Wertheim et al., 2004). Finally, given the extensive variability of virulence factors and mechanisms in S. aureus, our study has important implications for vaccine development. The data strongly suggest that the development of protective vaccines will require a very careful selection and combination of bacterial antigens.

Proteogenomics uncovers extreme heterogeneity in the Staphylococcus aureus
exoproteome due to genomic plasticity and variant gene regulation
67
Acknowledgements
We are indebted to J. Ziebuhr for critical comments on the manuscript and S. Holtfreter and S. Kozitskaya for assistance in some experiments. Financial support was provided by CEU (StaphDynamics, LSHM-CT-2006-019064; BaSysBio, LSHG-CT-2006-037469), BMBF (031U107A/-207A; 031U213B), DFG (GK212/3-00, SFB/TR34, FOR 585), and Top Institute Pharma (T4-213).

68
“Music is my religion”
-Johnny A. Hendrix-