Secondary Drug Resistance Mutation of TEM-1 -lactamase
description
Transcript of Secondary Drug Resistance Mutation of TEM-1 -lactamase
Secondary Drug Resistance Mutation of TEM-1 -lactamase
Amino acid sequence determinants of -lactamase structure and activity J. Mol. Biol. 258, 688-703 (1996)
A natural polymorphism in -lactamase is a global suppressor Proc. Natl. Acad. Sci. USA 94, 8801-8806 (1997)
A secondary drug resistance mutation of TEM-1 -lactamase that suppresses misfolding and aggregation Proc. Natl. Acad. Sci. USA 98, 283-288 (2001)
授課教授:楊孝德老師 指導教授:呂平江老師
報告學生:徐芝琪
Background
-lactam antibiotics inhibit the cross-linking transpeptidase and interfere with the synthesis of the bacterial cell wall.
TEM-1 -lactamase provides the major mechanism of plasmid-mediated -lactam resistance.
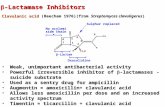
Clavulanic acid is the natural inhibitor of -lactamase.Natural variants of TEM-1 have more efficient at
catalyzing -lactam hydrolysis or more resistant to inhibitors.
Previous study in 1996 In a previous study, they performed saturation
mutagenesis in which each of the 263 codons of
the gene for TEM-1 -lactamase were randomized
by oligonucleotide-directed mutagenesis.
The essential residues are largely located in the
active site pocket or at buried positions in the
protein.
One of the critical residues that is located at a
buried position in the enzyme is Leu76. However,
buried residues are often tolerant of substitutions
of other hydrophobic residues.
Previous study in 1997
Introducing the gene for the L76N -lactamase into a mutS mismatch repair deficient strain.
Then the plasmid DNA was isolated and used to transform E. coli XL1-Blue.
The transformants were grown on agar plates containing 500 ug/ml ampicillin.
The DNA sequence of six revertants were determined: One mutant reverted codon 76 to Leu.
One mutant mutated codon 76 to Ile.
Four mutants converted codon 182 from Met to Thr.
‘‘Secondary’’ mutations compensate for defects?
Among the 90 TEM-1 natural isolates found to date,
substitutions at residues that directly affect substrate
binding and catalysis (R164, G238, and E104) or
inhibitor binding (M69 and R244) are repeatedly found.
M182T does not have a direct role in catalysis or
substrate/inhibitor binding and is never found alone.
In the 3D structure, Met182 is far away from the sites of
the coupled primary mutation so that any suppression
mechanism must operate at a distance.
Minimal inhibitory concentration ofampicillin and specific activities
•M182T causes a slight (20–30%) increase in sensitivity to ampicillin and comparable decrease in the -lactamase activity.•However, in combination with L76N, the M182T suppressor restores ampicillin resistance to near wild-type values and increases the -lactamase activity to 40% of the wild type.
Kinetic parameters of -lactamase catalysis of ampicillin hydrolysis
•The catalytic efficiency of L76N is 30% of wild type, but M182T by itself has no effect on kcat or Km values.•When M182T is combined with L76N, it causes a slight increase in activity against ampicillin, resulting in a double mutant with 53% the catalytic efficiency of the wild type.
When combined with L76N, the suppressor M182T largely restores the amount of -lactamase.
If the low expression of L76N is due to increased proteolysis, expressing it in SB646 strain should enhance its expression levels.
But the majority (67%) of L76N is in a catalytically inactive.
-lactamase express to the correctly folded, soluble, and enzymatically active form in M182T/L76N mutant.
Expression Levels of Mutant and Wild-Type -lactamase
206% 10715% +70%
CD spectroscopy: The final protein concen-tration
was 15 mM. For the wild-type protein, the first
transition (NI) has a mid-point at 0.8 M and the second (IU) at 2.3 M.
Fluorescence spectroscopy: The final protein concen-tration
was 0.6 mM (excited at 290 nm, and emission was monitored at 340 nm).
Fluorescence reveals only one transition corresponds to the first transition observed by CD.
Equilibrium denaturation of TEM-1 -lactamase
Thermodynamic parameters for the equilibrium transitions
The native protein is destabilized only in the double mutant.
Aggregation properties of wild-type and mutant -
lactamasesBecause the effects of these mutations cannot be
attributed to changes in stability or intrinsic catalytic activity, the remaining possibility is an effect on protein folding and/or aggregation.
The periplasmic extracts were denatured in GdnHCl and then diluted to allow refolding.
These results suggest that the L76N mutation promotes the accumulation of misfolded and aggregated -lactamase.
Conclusion
Because the majority of the double mutant (L76N/M182T) is soluble and active within the periplasm, the M182T mutation suppresses this enhanced aggregation and misfolding caused by the L76N mutation.
M182T may destroy an aggregation-prone site created by L76N or alter the folding mechanism to kinetically disfavor aggregation and misfolding.
-lactamase catalyzes the hydrolysis of -lactam ring
Most clinically important -lactamases employ a mechanism involving a nucleophilic serine residue and proceeding via a hydrolytically labile acyl-enzyme intermediate.
Back
Inhibition of serine -lactamase by clavulanic acid
It inhibits serine -lactamases through a complex mechanism involving sequential acylation, decarboxylation and loss of a four carbon fragment to give stable acyl-enzyme complexes.
Back