Resting State Networks in Subjects under Low and High ... · Resting State Networks in Subjects...
Transcript of Resting State Networks in Subjects under Low and High ... · Resting State Networks in Subjects...

Resting State Networks in Subjects under Low and High Sleep Pressure
1 University of Zurich, Institute of Pharmacology and Toxicology, Chronobiology and Sleep Research, Switzerland2 University of Zurich and ETH Zurich, Neuroscience Center Zurich, Switzerland3 ETH Zurich, Neural Control of Movement Lab, Department of Health Sciences and Technology, Switzerland4 Trinity College Dublin, Trinity College Institute of Neuroscience, Ireland
5 University of Zurich, University Clinics for Child and Adolescent Psychiatry, (UCCAP), Switzerland 6 Medical Faculty Mannheim/ Heidelberg University, Department of Child and Adoles-cent Psychiatry and Psychotherapy, Central Institute of Mental Health, Germany7 University of Zurich, Zurich Center for Integrative Human Physiology, Switzerland8 University of Zurich, Zurich Center for Interdisciplinary Sleep Research, Switzerland
SNF Sinergia Grant #136249
• Studies exploring resting state networks (RSNs) increa- sed, typically found 10 RSNs (focus on default mode net- work (DMN)).• Sleep studies reported - sleep stage dependency of DMN connectivity and - sleep pressure dependent changes of intra-network connectivity.
• Few studies examined RSNs with ASL (absolute measure of cerebral blood flow, CBF).
Thus, we conducted a RSN analysis on combined BOLD/CBF data, from subjects under low and high sleep pressure.
BACKGROUND
MATERIAL AND METHODS
• RSNs analysis of combined BOLD/CBF data (eyes open) was performed with the GIFT toolbox [1] (20 independent components; ICs)• 15 ICs were spatially correlated to the 28 RSNs described by Allen et al. [2] and grouped to networks• Statistical differences regarding spatial maps and FC were obtained with the MANCOVAN toolbox implemented in GIFT
REFERENCES[1] Group ICA of fMRI Toolbox http://mialab.mrn.org
[2] Allen et al. Front Syst Neurosci 2011[3] Perlis et al. J Sleep Res 1997
23:30 03:30 21:30
~18.6 h of extended wakefulness4 h of sleep (sleep lab)
RS recs
23:30 07:30 15:30/16:30
approx. 8 h of sleep (at home)
RS recs
ACKNOWLEDGMENTS CONTACTLaura TüshausUniversity of ZurichInstitute of Pharmacology and ToxicologyChronobiology and Sleep [email protected]
@
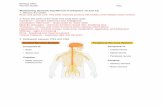
RESULTS Fifteen ICs highly correlated with RSN ICs described previously [2].Spatial maps did not differ be-tween groups.However, under high sleep pres-sure, FC between a sensorimotor IC and a visual IC was increased (p<0.05, FDR corr.).
STUDY PROTOCOL DATA PROCESSING
CONCLUSIONS
Sleep pressure changed functional connectivity (FC) between RSNs rather than altering their spatial extent. - Hyperarousal model of insomnia [3]: increased FC between primary sensory cortices and the supplementary motor area may contribute to sustained sensory processing, and thus prolonging sleep latency.- Increased FC may reflect the effort to stay awake in our high sleep pressure subjects.
Laura Tüshaus1,2,8, J. Balsters3,4, A. Schläpfer2,5, D. Brandeis2,5,6,7, P. Achermann1,2,7,8
Realign ASL images
Quantify CBF
Coregister CBF images with T1, normalize to MNI space
ASL
Smooth CBF images
Realign BOLD images
Coregister images with T1, normalize to MNI space
BOLD
Smooth images
Group 1
Group 2
~8.9 h of wakefulness
Figure 1a) Spatial map of IC11 (pink), highest correlation to the visual network of Allen et al. [2] (cyan).
b) Spatial map of IC15 (blue), highest correlation to the sen- sorimotor network of Allen et al. [2] (yellow).
Both ICs shown as t-maps thresholded at tc ≥ µc+ 4sc .
1)
a) b)
Travel grants:
Group 1, low sleep pressure Group 2, high sleep pressure
age/gender mean 24 ± 2.4 y, 10 f, 7 m mean 22.1 ± 2.4 y, 17 m
sleep normal restricted to 4 h
wake time ~8.9 ± 2.2 h ~18.6 ± 0.3 h
BOLD 2x 2 min eyes open/closed 2x 2 min eyes open/closed
EPI, TR/TE 1.96 s/30 ms,35 slices, voxels 3x3x3 mm3
EPI, TR/TE 1.96 s/30 ms,35 slices, voxels 3x3x3 mm3
ASL 5 min eyes open/closed 10 min eyes open
pCASL, TR/TE 4.4 s/20 ms,label duration 1.65 spost label delay 1.525 s,20 slices, voxels 3x3x7 mm3
pCASL, TR/TE 4.4 s/20 ms,label duration 1.65 spost label delay 1.525 s,20 slices, voxels 3x3x7 mm3