RESEARCH ARTICLE Open Access Repetitive part of …The information obtained in this study improves...
Transcript of RESEARCH ARTICLE Open Access Repetitive part of …The information obtained in this study improves...

RESEARCH ARTICLE Open Access
Repetitive part of the banana (Musa acuminata)genome investigated by low-depth454 sequencingEva Hřibovaacute1 Pavel Neumann2 Takashi Matsumoto3 Nicolas Roux4 Jiřiacute Macas2 Jaroslav Doležel1
Abstract
Background Bananas and plantains (Musa spp) are grown in more than a hundred tropical and subtropicalcountries and provide staple food for hundreds of millions of people They are seed-sterile crops propagatedclonally and this makes them vulnerable to a rapid spread of devastating diseases and at the same time hampersbreeding improved cultivars Although the socio-economic importance of bananas and plantains cannot beoverestimated they remain outside the focus of major research programs This slows down the study of nucleargenome and the development of molecular tools to facilitate banana improvement
Results In this work we report on the first thorough characterization of the repeat component of the banana(M acuminata cv lsquoCalcutta 4prime) genome Analysis of almost 100 Mb of sequence data (015times genome coverage)permitted partial sequence reconstruction and characterization of repetitive DNA making up about 30 of thegenome The results showed that the banana repeats are predominantly made of various types of Ty1copia andTy3gypsy retroelements representing 16 and 7 of the genome respectively On the other hand DNA transposonswere found to be rare In addition to new families of transposable elements two new satellite repeats werediscovered and found useful as cytogenetic markers To help in banana sequence annotation a specific Musarepeat database was created and its utility was demonstrated by analyzing the repeat composition of 62 genomicBAC clones
Conclusion A low-depth 454 sequencing of banana nuclear genome provided the largest amount of DNAsequence data available until now for Musa and permitted reconstruction of most of the major types of DNArepeats The information obtained in this study improves the knowledge of the long-range organization of bananachromosomes and provides sequence resources needed for repeat masking and annotation during the Musagenome sequencing project It also provides sequence data for isolation of DNA markers to be used in geneticdiversity studies and in marker-assisted selection
BackgroundBananas and plantains (Musa spp) are perennial giantherbs grown in humid tropical and subtropical regionsTheir annual production exceeds 100 million tons outof which almost 90 is targeted for local and nationalmarkets [1] Cultivated bananas are parthenocarpicseed-sterile vegetatively-propagated diploid triploid andtetraploid clones Most of them are hybrids between twodiploid (2n = 2x = 22) species M acuminata and
M balbisiana [2] with the A and B genomes respec-tively The production of bananas is threatened by manydiseases and pests but the clonal nature seed sterilityand the lack of knowledge on the origin of cultivatedclones hampers breeding of improved cultivars It isexpected that the use of molecular tools will speed upbanana germplasm improvement Sadly although thesocio-economic importance of bananas and plantainscannot be questioned Musa remains outside the focusof major research programs and must be considered anunder-researched cropThis situation is reflected by a limited knowledge of
the banana nuclear genome even though it is relatively
Correspondence hribovauebcascz1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech RepublicFull list of author information is available at the end of the article
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
copy 2010 Hřibovaacute et al licensee BioMed Central Ltd This is an Open Access article distributed under the terms of the Creative CommonsAttribution License (httpcreativecommonsorglicensesby20) which permits unrestricted use distribution and reproduction inany medium provided the original work is properly cited
small (1C ~ 600 Mbp) [34] It has been estimated thatabout 55 of the genome is made of various DNArepeats [5] but only a limited number of repetitiveDNA sequences has been characterized Valaacuterik et al(2002) described twelve Radka repeats [6] representingpartial sequences of various mobile elements and rRNAgenes Other characterized sequences included a Copia-like element [78] a species-specific element Brep-1[910] and a Ty3gypsy-like retrotransposon monkey[11] In order to identify more repeats Hřibovaacute et al [5]applied a low-Cot DNA isolation technique to charac-terize highly repetitive fractions of banana genome Animportant step forward in dissecting the Musa genomewas made by Cheung and Town (2007) who sequencedends of more than 6000 BAC (Bacterial Artificial Chro-mosome) clones [12] Moreover 62 BAC clones werecompletely sequenced through a Generation ChallengeProgramme funded project (GCP 2005-15) within thecontext of the Global Musa Genomics Consortium [13]Nevertheless even after these efforts the knowledge onthe repetitive part of Musa genome remains far fromcompleteRecent introduction of the next generation sequencing
methods [14] provided powerful tools to discover andcharacterize DNA repeats even in complex plant gen-omes For example Macas et al (2007) used the 454technology to characterize repetitive DNA in the nucleargenome of pea (Pisum sativum L) [15] Despite the rela-tively small proportion of sequenced DNA relative tothe whole genome (333 Mb or ~ 077 of the genome)the authors identified and characterized most types ofretrotransposons and discovered thirteen new families oftandemly organized repeats In a similar study Swami-nathan et al (2007) used the 454 system to sequence75 of the soybean genome [16]This study addresses the lack of knowledge on the
repetitive part of the banana genome by characterizingall major DNA repeats after massively parallel sequen-cing of genomic DNA of a diploid clone of M acumi-nata The experimental approach follows that of Macaset al [15] in which all-to-all similarity comparison of454 reads is performed to identify groups (clusters) ofoverlapping reads representing repetitive genomicsequences As the number of reads in individual clustersis proportional to genomic abundance of correspondingrepeats this information can be used for quantitativeanalysis of repetitive genome landscape In additionconsensus sequences of the repeated elements can beobtained by assembling the reads within clusters Wealso demonstrate how databases of 454 reads sortedaccording to the type of repeats can be used to identifyand classify repeats in BAC sequences Finally the largeset of sequences obtained in this study provides aunique source of molecular markers potentially useful in
genome mapping anchoring physical maps analyzinggenetic diversity and for phylogenetic studies
Results and DiscussionA diploid clone M acuminata cv lsquoCalcutta 4prime was cho-sen for sequencing as it has been used extensively as amodel genotype in previous molecular studies[561217-19] Moreover this clone is being used invarious banana breeding programs as a source of dis-eases resistance [2021] A sequencing run of nuclearDNA on the GS FLX platform (454 Life SciencesRoche) resulted in 477699 reads with average lengthof 206 bp providing a total of 98538911 bp ofsequence data Considering genome size of lsquoCalcutta 4prime(1C = 623 Mbp) [4] this represents 157 of the gen-ome The sequencing reads were clustered based ontheir similarity and all clusters containing at least20 reads (roughly representing 001 of the genome)were further investigated
LTR-retrotransposonsThe most abundant DNA sequences found in thebanana genome were LTR-retrotransposons Out ofthem Ty1copia represented more than 16 of the gen-ome while the Ty3gypsy elements represented about 7of the genome (Figure 1) This is an interesting observa-tion as the available data from other sequencing projectsindicate prevalence of Ty3gypsy retrotransposons inplant nuclear genomes [1522-24] In order to get insightinto the diversity of banana LTR-retrotransposons weperformed phylogenetic analysis based on a comparisonof their reverse transcriptase domains This workrevealed that while the more abundant Ty1copia-likeelements were represented by four distinct evolutionarylineages (Figure 2A) vast majority of Ty3gypsy-like ele-ments belonged to a single evolutionary lineage of chro-moviruses [25] (Figure 2B)About 74 of identified Ty1copia sequences belonged
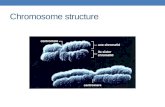
to the SIREMaximus lineage [2627] representingalmost 13 of the genome The remaining Ty1copiaelements belonging to Angela Tnt1 and Hopscotchlineages [27-29] represented only about 30 10 and02 of the genome respectively Interestingly fluores-cence in situ hybridization (FISH) on mitotic chromo-somes revealed that elements from distinct evolutionarylineages have different patterns of genomic distributionThe elements from the SIREMaximus and Angelalineages were concentrated in several discrete clusterson all chromosomes (Figures 3D E) and the elementsfrom the Tnt-1 lineage gave only weak signals preferen-tially localized in distal parts of mitotic chromosomes(Figure 3F) Elements belonging to the Hopscotch line-age were not tested for the distribution because of theirvery low proportion in the genome
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 2 of 10
Ty3gypsy-like retrotransposons showed relatively lowdegree of phylogenetic diversity and most of thembelonged to the lineage of chromoviruses This singlelineage comprised about 87 of Ty3gypsy elementsidentified in this study thus greatly outnumbering ele-ments from the Tat lineage which included all otherTy3gypsy elements identified in the banana genomeThe chromoviral sequences could be classified into fourclades Galadriel Tekay Reina and CRM [30-32] Themost abundant chromoviral clade was Reina whichinvolved more than half of all chromoviral sequencesmaking up about 4 of the banana genome Many ele-ments belonging to this clade appeared to be non-autonomous as they lacked parts of RT-coding domain(data not shown) Members of the Tekay clade werefound to be the second most abundant group of chro-moviruses reaching about 2 of the genome Sequencesfrom the Galadriel clade corresponded to the retrotran-sposon monkey which has been identified earlier in thebanana genome [11] The consensus sequences of themonkey retrotransposon assembled from our 454 data asa 5880 bp fragment showed 95 similarity to the mon-key element described by Balint-Kurti et al (2000) [11]Previous estimates of the copy number using slot-blotanalysis indicated that monkey constituted about 02 -05 of the M acuminata genome [11] and are on linewith our estimates based on the proportion of monkey-derived sequences in 454 reads (Additional file 1)Although the monkey was supposed to be the mostabundant repetitive element in banana [5] our datashowed that several other families of retroelementsaccount for much larger parts of the genome The CRMclade sequences occupied a similar fraction of the gen-ome as those from the Galadriel clade Although beingmembers of the same evolutionary lineage banana chro-moviruses from distinct clades partly differed in their
chromosomal distribution Contrary to monkey whichpreferentially localized in secondary constrictions [11]members of other clades occupied mostly pericentro-meric regions and some additional loci in distal parts ofall chromosomes (Figures 3G H)
Non-LTR retrotransposons and DNA transposonsCompared to LTR-retrotransposons non-LTR retrotran-sposons and DNA transposons were found relativelyrare (Additional file 1) Within the clusters that repre-sented at least 001 of the genome only one cluster ofLINE sequences [33] and two clusters of DNA transpo-sons were identified The LINE elements were estimatedto constitute about 1 of the banana genome FISHwith a probe derived from reverse transcriptase domainof a LINE-like element resulted in dot signals in centro-meric regions on all chromosomes (Figure 3I) DNAtransposons identified in this work included elementsthat showed similarity to transposons belonging to thehAT superfamily [34] FISH with a probe derived fromhAT-related element failed to give visible signals mostprobably due to relatively small copy number The lowabundance of LINEs and DNA transposons seems to betypical for plant genomes and similar abundances wereobserved for example in rice grape and maize genomes[232435]
45S and 5S rDNAClusters containing 45S rDNA represented 112 of thegenome and the 45S rDNA sequence region was recon-structed as a 7553 bp fragment that included completesequence of the 18S-58S-26S rRNA locus surroundedby parts of intergenic spacer (IGS) Moreover based onsimilarity searches to BAC clone MA4_01C21 fromM acuminata which was sequenced within the contextof the Global Musa Genomics Consortium [13] and
Figure 1 Genome proportion of major groups of repetitive sequences identified in banana 454 data
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 3 of 10
which carries 45S rDNA units another cluster contain-ing IGS-like sequence was identified in the 454 dataIn contrast to Balint-Kurti et al (2000) whose results
obtained after FISH with mitotic chromosomes indi-cated insertion of a part of monkey into 45S rDNA [11]our 454 data suggests that monkey is not frequentlyassociated with the 18S-58S-26S rRNA gene copies
A plausible explanation for this discrepancy is that theinsertion is adjacent to the 45S rDNA locus In fact thespatial resolution of FISH on mitotic chromosomes isnot sufficient to discriminate two loci closer than 5 - 10Mbp [3637] A close vicinity of the monkey fragment to45S rDNA is supported by the sequence data of theBAC MA4_01C21 comprising 45S rDNA in which a
Figure 2 Phylogenetic analysis of Musa retrotransposons based on RT sequences Unrooted phylogenetic trees of Ty1copia elements (A)and Ty3gypsy elements (B) Names of the contigs assembled from 454 reads are printed in purple Classification of the Ty3gypsy lineages andchromoviral clades was done according to [30-32] Major lineages of Ty1copia elements were named according to a selected representative ofeach group
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 4 of 10
15kb fragment of monkey was identified However thefragment was not inserted in the 45SrDNA or IGSsequences The fact that this BAC comprises a chromo-virus element from the Tekay lineage and the SIRE1Maximus lineage indicates that the BAC MA4_01C21actually encompasses a border of the 45S rDNA locusand flanking genomic sequences characterized bysequence-heterogeneity and insertion of various mobileelementsSimilar to the 45S rRNA gene cluster our 454
sequence data enabled reconstruction of the entire cod-ing part of the 5S rRNA gene and its non-transcribedspacer The 5S rDNA was found to represent 038 ofthe banana nuclear genome Teo et al [38] identified aTy1copia-like element in the 5S rDNA spacer in severalbanana species The analysis of retrotransposon proteincoding domains in our data confirmed that the 5SrDNA spacer contained a part of the reverse transcrip-tase of the Tnt1-like element
Tandem organized repeatsRepeat reconstruction from the 454 data led to discov-ery of two new tandemly organized repeats One ofthem (CL33) consists of ~130 bp monomer while theCL18 repeat is characterized by ~2 kb monomer unitFISH on mitotic chromosomes revealed clusters of sig-nals in the subtelomeric regions of one pair of chromo-somes (satellite CL18) and weak signals in telomericregion on two pairs of chromosomes (satellite CL33)(Figures 3A B C) Southern hybridization resulted in aladder-like pattern typical for tandemly organized repeti-tive units for repeat CL33 only (not shown) The repeatCL18 gave a weak smear with a few visible bands mostlikely due to partially dispersed distribution andor poorconservation of the monomer length A rather low copynumber of CL33 andor long repetitive unit of satelliteCL18 may explain why these repeats were not identifiedin previous studies [56] In general the absence ofmore abundant tandem repeats in the banana genomemay be related to its relatively small size Satellite DNAis a typical component of subtelomeric and centromericchromosome regions in various plant species but theyoften form blocks of repeats in interstitial regions[1539-42] The results of this study as well as our ear-lier observations [56] indicate that typical centromericsatellite DNA is absent in the banana genome and thatthe centromeric regions are likely to be made of varioustypes of retrotransposons
Identification of DNA markersFollowing the thorough characterization of banana repe-titive DNA we screened the 454 sequences for the pre-sence of loci potentially suitable for use as DNAmarkers We focused on identification of simple
sequence repeats (SSRs) and sites of insertions of trans-posable elements (ISBP - Insertion Site Based Poly-morphism) [43] In total 27946 of 454 reads containingSSRs were identified with repeat units ranging from 2 to10 bp The most abundant motifs were dinucleotidesTA and GA and trinucleotides GAA (Figure 4) Morethan 11000 reads were identified to contain potentialISBP sites most of them carrying insertions of retro-transposons into unknown low-copy sequences Data-bases containing 454 reads carrying SSR sequences andpotential ISBPs were established and made publiclyavailable on our website [44]
Repeat identification in sequenced DNA clonesAs the next step in utilizing 454 data we took advantageof the read clustering during repeat analysis and createddatabases of sequence reads sorted according to theirrepeat of origin These databases were then utilized forsimilarity-based repeat detection and classification ingenomic BAC clone sequences implemented at thePROFREP server [45] The analysis was performed for49 BAC clones from M acuminata cv lsquoCalcutta 4prime andfor 12 BAC clones from M balbisiana cv lsquoPisang Klu-tug Wulungrsquo [14] The clones were sequenced as part ofthe Generation Challenge Program supported project(GCP-2005-15) and were selected based on the presenceof resistance gene homologs and other gene-likesequences Indeed out of the 49 M acuminata BACclones only 9 clones were highly repetitive 15 BACclones contained a single copy sequence with a largerepetitive region and the remaining BAC clones com-prised single copy sequence without any detectable repe-titive DNA andor carried a very short repetitive region(Figure 5) Out of the 12 BAC clones from M balbisi-ana two were single copy while the remaining 10 BACclones carried low copy sequences mixed with largerepetitive regions The repetitive profiles of all 62 BACclones are available as supplementary data (Additionalfile 2)
ConclusionsThis work represents a major advance in the analysis ofthe nuclear genome organization in banana an impor-tant staple and cash crop The application of low-depth454 sequencing provided until now the largest amountof DNA sequence data and enabled a detailed analysisof repetitive components of its nuclear genome Allmajor types of DNA repeats were characterized andMusa DNA repeats databases were established The ana-lysis of genomic distribution of selected repeats providednew data on long-range molecular organization ofbanana chromosomes and a large number of loci poten-tially useful as DNA markers were identified Theimproved knowledge and resources generated in this
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 5 of 10
Figure 3 Genomic distribution of different types of DNA repeats Mitotic metaphase spreads of M acuminata cv lsquoCalcutta 4rsquo (2n = 22) afterFISH with probes for various repeats The chromosomes were counterstained with DAPI (blue) Bar = 5 μm (A) Tandem repeat CL18 (greensignal) formed a cluster on one pair of chromosomes (long arrows) (B) Tandem repeat CL33 (red signal) localized on two pairs of chromosomes(long arrows) (C) Simultaneous hybridization of probes for CL18 (green signal) and CL33 (red signal long arrows) revealed co-localization of bothsatellites on one pair of chromosomes (short arrows) (D) Two metaphase plates after FISH with a probe for CL1SCL2Contig1080 - bananaretrotransposon belonging to SIREMaximus lineage (green) Uneven genomic distribution with clusters dispersed on all chromosomes isobvious (E) Similar genomic distribution was found for banana retroelement related to the Angela lineage (CL2Contig49) (F) Bananaretrotransposon belonging to Tnt1 lineage (CL10Contig16 red color) gave weak signals preferentially localized in distal parts of chromosomes(long arrows) (G) The most abundant type of Ty3gypsy-like element of the Reina lineage (CL1SCL5Contig891) localized preferentially tocentromeric or peri-centromeric regions of all chromosomes (green signals) (H) Also the Ty3gypsy-like element related to Tekay evolutionarylineage (CL4Contig82) clustered in centromeric or peri-centromeric regions of all chromosomes (I) A probe derived from LINE element(CL1SCL8Contig452) localized in the centromeric regions of all chromosomes (green signals)
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 6 of 10
study will be useful in annotating the banana genomesequence in the analysis of the evolution of the Musagenome and for the study of dynamics of DNA repeatsover evolutionary time scale as well as to isolate DNAmarkers for use in genetic diversity studies and in mar-ker-assisted selection
Methods454 sequencingIn vitro rooted plants of M acuminata cv lsquoCalcutta 4prime(ITC 0249) were obtained from the International TransitCentre (Bioversity International Global Musa Genebankhosted by the Katholieke Universiteit Leuven Belgium)and grown in a greenhouse DNA for sequencing wasprepared from nuclei isolated from healthy young leaftissues according to Zhang et al (1995) [46] Isolatednuclei were incubated with 40 mM EDTA 02 SDSand 025 μgμl proteinase K for 5 hours at 37degC andDNA was purified by phenolchloroform precipitationThe 454 sequencing was performed at the ArizonaGenomics Institute (Tucson USA) using 454 LifeSciencesRoche FLX instrument All sequence informa-tion generated in this study are available on our website[44] and was submitted to the National Center for Bio-technology Information short read archive under acces-sion numbers SRR057410 and SRR057411
Data analysisFollowing a removal of linkerprimer contaminationsand artificially duplicated reads the remaining 477699reads (average length of 206 nucleotides) were used forrepeat analysis The analysis was performed as describedby Macas et al (2007) [15] employing TGICL [47] anda set of custom-made BioPerl scripts for similarity-basedclustering and assembly of reads The clustering para-meters used by a tclust program (part of TGICL) were
set to consider pairwise similarity of two reads signifi-cant if it involved an overlap of at least 150 nucleotideswith 90 or better similarity representing at least 55and 70 of the length of longer and shorter read respec-tively (OVL = 150 PID = 90 LCOV = 55 SCOV = 70)The reads within individual clusters were assembledinto contigs using TGICL run with the -O lsquo-p 80 -o 40primeparameters specifying overlap percent identity andminimal length cutoff for cap3 assembler Repeat typeidentification was done using blastn and blastx [48]sequence-similarity searches of assembled contigsagainst GenBank and by detection of conserved proteindomains using RPS-BLAST [49] Tandem repeats withincontig sequences were identified using dotter [50] Theclassification of LTR retrotransposons into distinctlineages and clades was done using phylogenetic ana-lyses of their RT sequences [15] Alignment of RTsequences was carried out with ClustalX [51] and thephylogenetic trees were calculated using neighbour-join-ing method The trees were drawn and edited using theFigTree programMicrosatellite sequences were identified using Tandem
Repeats Finder [52] and TRAP [53] programs while aBioPerl script was used to identify ISBP loci [54] Identi-fication and classification of repetitive sequences withinBAC clones was done via PROFREP web server [45] uti-lizing repeat-specific databases of 454 reads prepared inthis study The server performs BLAST-based searchesagainst databases of whole-genome or repeat-specific454 reads and generates plots of similarity hits along thequery sequence (number of hits is proportional to copynumber of the query in the genome)
Preparation of probes for cytogenetic mappingPrimers specific for tandem repeats (Additional file 3)were designed from sequence contigs that carry tandem
Figure 4 Major groups of microsatellite DNA sequences identified in banana 454 reads
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 7 of 10
organized repetitive units Labeled probes were preparedby PCR on M acuminata rsquoCalcutta 4prime genomic DNAwith biotin- and digoxigenin-labeled nucleotides ThePCR premix contained 1times PCR buffer 1 mM MgCl2 02mM dNTPs 02 μM primers 05 U Taq polymerase(Finnzymes) and 10 - 15 ng template DNA PCR reac-tion was performed as follows initial denaturation of3 min at 94degC followed by 30 cycles of 1 min at 94degC50 s at 57degC and 50 s at 72degC and final extension step5 min at 72degCSpecific primers were also designed for reverse tran-
scriptase (RT) domains of different retroelements(Additional file 3) In the first step the RT domainswere amplified using PCR with a mix containing 1timesPCR buffer 15 mM MgCl2 02 mM dNTPs 02 μMprimers 05 U Taq polymerase (Finnzymes) and 10 -15 ng template DNA PCR products were checked bygel-electrophoresis cleaned up using paramagneticbeads Agencourt Ampure (Beckman Coulter) clonedinto TOPO vector (Invitrogen) and transformed intoelectro-competent E coli cells 48 recombinant clonesfor each retroelement were PCR amplified using M13primers and separated on the gel electrophoresisClones for each RT domain were than cleaned upusing ExoSAP-IT (USB Corporation) and used forSanger sequencing to verify presence of specific RTdomains in the clones The PCR products weresequenced with BigDye Terminator 31 Cycle Sequen-cing Kit (Applied Biosystems) on ABI 3730xl DNAanalyzer (Applied Biosystems) The nucleotidesequences were analyzed and edited using the StadenPackage [55] and searched for similarity with the cor-responding 454 contigs using BLAST [48] Clones withthe highest similarity to reconstructed contigs werePCR amplified with biotin- and digoxigenin-labelednucleotides and used as probes for fluorescence in situhybridization Selected clones used as probes showedat least 98 similarity with the corresponding 454sequence
Fluorescence in situ hybridization (FISH)FISH was done on mitotic metaphase spreads preparedfrom meristem root tip cells as described by Doleželovaacuteet al (1998) [56] The hybridization mixture consistedof 40 formamide 10 dextran sulfate in 1 times SSC anda 1 μgml labeled probe The mixture was added ontoslides and denatured at 80degC for 4 min The hybridiza-tion was carried out at 37degC overnight The sites ofprobe hybridization were detected using anti-digoxi-genin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories) and the chromosomes werecounterstained with DAPI The slides were examinedwith Olympus AX70 fluorescence microscope
Figure 5 Examples of repeat identification in M acuminataBAC clones Nucleotide sequences of BAC inserts are representedon X axis The plots represent genomic copy numbers of individualinsert regions calculated from numbers of similarity hits to 454 readdatabases The plot colors correspond to different types of therepeats (A) A lsquolow-copyrsquo BAC clone showing absence of repeatsalong its entire length (B) A BAC clone with relatively long stretchesof repetitive DNA (C) A highly repetitive BAC clone with varioustypes of repeats
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 8 of 10
(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

small (1C ~ 600 Mbp) [34] It has been estimated thatabout 55 of the genome is made of various DNArepeats [5] but only a limited number of repetitiveDNA sequences has been characterized Valaacuterik et al(2002) described twelve Radka repeats [6] representingpartial sequences of various mobile elements and rRNAgenes Other characterized sequences included a Copia-like element [78] a species-specific element Brep-1[910] and a Ty3gypsy-like retrotransposon monkey[11] In order to identify more repeats Hřibovaacute et al [5]applied a low-Cot DNA isolation technique to charac-terize highly repetitive fractions of banana genome Animportant step forward in dissecting the Musa genomewas made by Cheung and Town (2007) who sequencedends of more than 6000 BAC (Bacterial Artificial Chro-mosome) clones [12] Moreover 62 BAC clones werecompletely sequenced through a Generation ChallengeProgramme funded project (GCP 2005-15) within thecontext of the Global Musa Genomics Consortium [13]Nevertheless even after these efforts the knowledge onthe repetitive part of Musa genome remains far fromcompleteRecent introduction of the next generation sequencing
methods [14] provided powerful tools to discover andcharacterize DNA repeats even in complex plant gen-omes For example Macas et al (2007) used the 454technology to characterize repetitive DNA in the nucleargenome of pea (Pisum sativum L) [15] Despite the rela-tively small proportion of sequenced DNA relative tothe whole genome (333 Mb or ~ 077 of the genome)the authors identified and characterized most types ofretrotransposons and discovered thirteen new families oftandemly organized repeats In a similar study Swami-nathan et al (2007) used the 454 system to sequence75 of the soybean genome [16]This study addresses the lack of knowledge on the
repetitive part of the banana genome by characterizingall major DNA repeats after massively parallel sequen-cing of genomic DNA of a diploid clone of M acumi-nata The experimental approach follows that of Macaset al [15] in which all-to-all similarity comparison of454 reads is performed to identify groups (clusters) ofoverlapping reads representing repetitive genomicsequences As the number of reads in individual clustersis proportional to genomic abundance of correspondingrepeats this information can be used for quantitativeanalysis of repetitive genome landscape In additionconsensus sequences of the repeated elements can beobtained by assembling the reads within clusters Wealso demonstrate how databases of 454 reads sortedaccording to the type of repeats can be used to identifyand classify repeats in BAC sequences Finally the largeset of sequences obtained in this study provides aunique source of molecular markers potentially useful in
genome mapping anchoring physical maps analyzinggenetic diversity and for phylogenetic studies
Results and DiscussionA diploid clone M acuminata cv lsquoCalcutta 4prime was cho-sen for sequencing as it has been used extensively as amodel genotype in previous molecular studies[561217-19] Moreover this clone is being used invarious banana breeding programs as a source of dis-eases resistance [2021] A sequencing run of nuclearDNA on the GS FLX platform (454 Life SciencesRoche) resulted in 477699 reads with average lengthof 206 bp providing a total of 98538911 bp ofsequence data Considering genome size of lsquoCalcutta 4prime(1C = 623 Mbp) [4] this represents 157 of the gen-ome The sequencing reads were clustered based ontheir similarity and all clusters containing at least20 reads (roughly representing 001 of the genome)were further investigated
LTR-retrotransposonsThe most abundant DNA sequences found in thebanana genome were LTR-retrotransposons Out ofthem Ty1copia represented more than 16 of the gen-ome while the Ty3gypsy elements represented about 7of the genome (Figure 1) This is an interesting observa-tion as the available data from other sequencing projectsindicate prevalence of Ty3gypsy retrotransposons inplant nuclear genomes [1522-24] In order to get insightinto the diversity of banana LTR-retrotransposons weperformed phylogenetic analysis based on a comparisonof their reverse transcriptase domains This workrevealed that while the more abundant Ty1copia-likeelements were represented by four distinct evolutionarylineages (Figure 2A) vast majority of Ty3gypsy-like ele-ments belonged to a single evolutionary lineage of chro-moviruses [25] (Figure 2B)About 74 of identified Ty1copia sequences belonged
to the SIREMaximus lineage [2627] representingalmost 13 of the genome The remaining Ty1copiaelements belonging to Angela Tnt1 and Hopscotchlineages [27-29] represented only about 30 10 and02 of the genome respectively Interestingly fluores-cence in situ hybridization (FISH) on mitotic chromo-somes revealed that elements from distinct evolutionarylineages have different patterns of genomic distributionThe elements from the SIREMaximus and Angelalineages were concentrated in several discrete clusterson all chromosomes (Figures 3D E) and the elementsfrom the Tnt-1 lineage gave only weak signals preferen-tially localized in distal parts of mitotic chromosomes(Figure 3F) Elements belonging to the Hopscotch line-age were not tested for the distribution because of theirvery low proportion in the genome
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 2 of 10
Ty3gypsy-like retrotransposons showed relatively lowdegree of phylogenetic diversity and most of thembelonged to the lineage of chromoviruses This singlelineage comprised about 87 of Ty3gypsy elementsidentified in this study thus greatly outnumbering ele-ments from the Tat lineage which included all otherTy3gypsy elements identified in the banana genomeThe chromoviral sequences could be classified into fourclades Galadriel Tekay Reina and CRM [30-32] Themost abundant chromoviral clade was Reina whichinvolved more than half of all chromoviral sequencesmaking up about 4 of the banana genome Many ele-ments belonging to this clade appeared to be non-autonomous as they lacked parts of RT-coding domain(data not shown) Members of the Tekay clade werefound to be the second most abundant group of chro-moviruses reaching about 2 of the genome Sequencesfrom the Galadriel clade corresponded to the retrotran-sposon monkey which has been identified earlier in thebanana genome [11] The consensus sequences of themonkey retrotransposon assembled from our 454 data asa 5880 bp fragment showed 95 similarity to the mon-key element described by Balint-Kurti et al (2000) [11]Previous estimates of the copy number using slot-blotanalysis indicated that monkey constituted about 02 -05 of the M acuminata genome [11] and are on linewith our estimates based on the proportion of monkey-derived sequences in 454 reads (Additional file 1)Although the monkey was supposed to be the mostabundant repetitive element in banana [5] our datashowed that several other families of retroelementsaccount for much larger parts of the genome The CRMclade sequences occupied a similar fraction of the gen-ome as those from the Galadriel clade Although beingmembers of the same evolutionary lineage banana chro-moviruses from distinct clades partly differed in their
chromosomal distribution Contrary to monkey whichpreferentially localized in secondary constrictions [11]members of other clades occupied mostly pericentro-meric regions and some additional loci in distal parts ofall chromosomes (Figures 3G H)
Non-LTR retrotransposons and DNA transposonsCompared to LTR-retrotransposons non-LTR retrotran-sposons and DNA transposons were found relativelyrare (Additional file 1) Within the clusters that repre-sented at least 001 of the genome only one cluster ofLINE sequences [33] and two clusters of DNA transpo-sons were identified The LINE elements were estimatedto constitute about 1 of the banana genome FISHwith a probe derived from reverse transcriptase domainof a LINE-like element resulted in dot signals in centro-meric regions on all chromosomes (Figure 3I) DNAtransposons identified in this work included elementsthat showed similarity to transposons belonging to thehAT superfamily [34] FISH with a probe derived fromhAT-related element failed to give visible signals mostprobably due to relatively small copy number The lowabundance of LINEs and DNA transposons seems to betypical for plant genomes and similar abundances wereobserved for example in rice grape and maize genomes[232435]
45S and 5S rDNAClusters containing 45S rDNA represented 112 of thegenome and the 45S rDNA sequence region was recon-structed as a 7553 bp fragment that included completesequence of the 18S-58S-26S rRNA locus surroundedby parts of intergenic spacer (IGS) Moreover based onsimilarity searches to BAC clone MA4_01C21 fromM acuminata which was sequenced within the contextof the Global Musa Genomics Consortium [13] and
Figure 1 Genome proportion of major groups of repetitive sequences identified in banana 454 data
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 3 of 10
which carries 45S rDNA units another cluster contain-ing IGS-like sequence was identified in the 454 dataIn contrast to Balint-Kurti et al (2000) whose results
obtained after FISH with mitotic chromosomes indi-cated insertion of a part of monkey into 45S rDNA [11]our 454 data suggests that monkey is not frequentlyassociated with the 18S-58S-26S rRNA gene copies
A plausible explanation for this discrepancy is that theinsertion is adjacent to the 45S rDNA locus In fact thespatial resolution of FISH on mitotic chromosomes isnot sufficient to discriminate two loci closer than 5 - 10Mbp [3637] A close vicinity of the monkey fragment to45S rDNA is supported by the sequence data of theBAC MA4_01C21 comprising 45S rDNA in which a
Figure 2 Phylogenetic analysis of Musa retrotransposons based on RT sequences Unrooted phylogenetic trees of Ty1copia elements (A)and Ty3gypsy elements (B) Names of the contigs assembled from 454 reads are printed in purple Classification of the Ty3gypsy lineages andchromoviral clades was done according to [30-32] Major lineages of Ty1copia elements were named according to a selected representative ofeach group
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 4 of 10
15kb fragment of monkey was identified However thefragment was not inserted in the 45SrDNA or IGSsequences The fact that this BAC comprises a chromo-virus element from the Tekay lineage and the SIRE1Maximus lineage indicates that the BAC MA4_01C21actually encompasses a border of the 45S rDNA locusand flanking genomic sequences characterized bysequence-heterogeneity and insertion of various mobileelementsSimilar to the 45S rRNA gene cluster our 454
sequence data enabled reconstruction of the entire cod-ing part of the 5S rRNA gene and its non-transcribedspacer The 5S rDNA was found to represent 038 ofthe banana nuclear genome Teo et al [38] identified aTy1copia-like element in the 5S rDNA spacer in severalbanana species The analysis of retrotransposon proteincoding domains in our data confirmed that the 5SrDNA spacer contained a part of the reverse transcrip-tase of the Tnt1-like element
Tandem organized repeatsRepeat reconstruction from the 454 data led to discov-ery of two new tandemly organized repeats One ofthem (CL33) consists of ~130 bp monomer while theCL18 repeat is characterized by ~2 kb monomer unitFISH on mitotic chromosomes revealed clusters of sig-nals in the subtelomeric regions of one pair of chromo-somes (satellite CL18) and weak signals in telomericregion on two pairs of chromosomes (satellite CL33)(Figures 3A B C) Southern hybridization resulted in aladder-like pattern typical for tandemly organized repeti-tive units for repeat CL33 only (not shown) The repeatCL18 gave a weak smear with a few visible bands mostlikely due to partially dispersed distribution andor poorconservation of the monomer length A rather low copynumber of CL33 andor long repetitive unit of satelliteCL18 may explain why these repeats were not identifiedin previous studies [56] In general the absence ofmore abundant tandem repeats in the banana genomemay be related to its relatively small size Satellite DNAis a typical component of subtelomeric and centromericchromosome regions in various plant species but theyoften form blocks of repeats in interstitial regions[1539-42] The results of this study as well as our ear-lier observations [56] indicate that typical centromericsatellite DNA is absent in the banana genome and thatthe centromeric regions are likely to be made of varioustypes of retrotransposons
Identification of DNA markersFollowing the thorough characterization of banana repe-titive DNA we screened the 454 sequences for the pre-sence of loci potentially suitable for use as DNAmarkers We focused on identification of simple
sequence repeats (SSRs) and sites of insertions of trans-posable elements (ISBP - Insertion Site Based Poly-morphism) [43] In total 27946 of 454 reads containingSSRs were identified with repeat units ranging from 2 to10 bp The most abundant motifs were dinucleotidesTA and GA and trinucleotides GAA (Figure 4) Morethan 11000 reads were identified to contain potentialISBP sites most of them carrying insertions of retro-transposons into unknown low-copy sequences Data-bases containing 454 reads carrying SSR sequences andpotential ISBPs were established and made publiclyavailable on our website [44]
Repeat identification in sequenced DNA clonesAs the next step in utilizing 454 data we took advantageof the read clustering during repeat analysis and createddatabases of sequence reads sorted according to theirrepeat of origin These databases were then utilized forsimilarity-based repeat detection and classification ingenomic BAC clone sequences implemented at thePROFREP server [45] The analysis was performed for49 BAC clones from M acuminata cv lsquoCalcutta 4prime andfor 12 BAC clones from M balbisiana cv lsquoPisang Klu-tug Wulungrsquo [14] The clones were sequenced as part ofthe Generation Challenge Program supported project(GCP-2005-15) and were selected based on the presenceof resistance gene homologs and other gene-likesequences Indeed out of the 49 M acuminata BACclones only 9 clones were highly repetitive 15 BACclones contained a single copy sequence with a largerepetitive region and the remaining BAC clones com-prised single copy sequence without any detectable repe-titive DNA andor carried a very short repetitive region(Figure 5) Out of the 12 BAC clones from M balbisi-ana two were single copy while the remaining 10 BACclones carried low copy sequences mixed with largerepetitive regions The repetitive profiles of all 62 BACclones are available as supplementary data (Additionalfile 2)
ConclusionsThis work represents a major advance in the analysis ofthe nuclear genome organization in banana an impor-tant staple and cash crop The application of low-depth454 sequencing provided until now the largest amountof DNA sequence data and enabled a detailed analysisof repetitive components of its nuclear genome Allmajor types of DNA repeats were characterized andMusa DNA repeats databases were established The ana-lysis of genomic distribution of selected repeats providednew data on long-range molecular organization ofbanana chromosomes and a large number of loci poten-tially useful as DNA markers were identified Theimproved knowledge and resources generated in this
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 5 of 10
Figure 3 Genomic distribution of different types of DNA repeats Mitotic metaphase spreads of M acuminata cv lsquoCalcutta 4rsquo (2n = 22) afterFISH with probes for various repeats The chromosomes were counterstained with DAPI (blue) Bar = 5 μm (A) Tandem repeat CL18 (greensignal) formed a cluster on one pair of chromosomes (long arrows) (B) Tandem repeat CL33 (red signal) localized on two pairs of chromosomes(long arrows) (C) Simultaneous hybridization of probes for CL18 (green signal) and CL33 (red signal long arrows) revealed co-localization of bothsatellites on one pair of chromosomes (short arrows) (D) Two metaphase plates after FISH with a probe for CL1SCL2Contig1080 - bananaretrotransposon belonging to SIREMaximus lineage (green) Uneven genomic distribution with clusters dispersed on all chromosomes isobvious (E) Similar genomic distribution was found for banana retroelement related to the Angela lineage (CL2Contig49) (F) Bananaretrotransposon belonging to Tnt1 lineage (CL10Contig16 red color) gave weak signals preferentially localized in distal parts of chromosomes(long arrows) (G) The most abundant type of Ty3gypsy-like element of the Reina lineage (CL1SCL5Contig891) localized preferentially tocentromeric or peri-centromeric regions of all chromosomes (green signals) (H) Also the Ty3gypsy-like element related to Tekay evolutionarylineage (CL4Contig82) clustered in centromeric or peri-centromeric regions of all chromosomes (I) A probe derived from LINE element(CL1SCL8Contig452) localized in the centromeric regions of all chromosomes (green signals)
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 6 of 10
study will be useful in annotating the banana genomesequence in the analysis of the evolution of the Musagenome and for the study of dynamics of DNA repeatsover evolutionary time scale as well as to isolate DNAmarkers for use in genetic diversity studies and in mar-ker-assisted selection
Methods454 sequencingIn vitro rooted plants of M acuminata cv lsquoCalcutta 4prime(ITC 0249) were obtained from the International TransitCentre (Bioversity International Global Musa Genebankhosted by the Katholieke Universiteit Leuven Belgium)and grown in a greenhouse DNA for sequencing wasprepared from nuclei isolated from healthy young leaftissues according to Zhang et al (1995) [46] Isolatednuclei were incubated with 40 mM EDTA 02 SDSand 025 μgμl proteinase K for 5 hours at 37degC andDNA was purified by phenolchloroform precipitationThe 454 sequencing was performed at the ArizonaGenomics Institute (Tucson USA) using 454 LifeSciencesRoche FLX instrument All sequence informa-tion generated in this study are available on our website[44] and was submitted to the National Center for Bio-technology Information short read archive under acces-sion numbers SRR057410 and SRR057411
Data analysisFollowing a removal of linkerprimer contaminationsand artificially duplicated reads the remaining 477699reads (average length of 206 nucleotides) were used forrepeat analysis The analysis was performed as describedby Macas et al (2007) [15] employing TGICL [47] anda set of custom-made BioPerl scripts for similarity-basedclustering and assembly of reads The clustering para-meters used by a tclust program (part of TGICL) were
set to consider pairwise similarity of two reads signifi-cant if it involved an overlap of at least 150 nucleotideswith 90 or better similarity representing at least 55and 70 of the length of longer and shorter read respec-tively (OVL = 150 PID = 90 LCOV = 55 SCOV = 70)The reads within individual clusters were assembledinto contigs using TGICL run with the -O lsquo-p 80 -o 40primeparameters specifying overlap percent identity andminimal length cutoff for cap3 assembler Repeat typeidentification was done using blastn and blastx [48]sequence-similarity searches of assembled contigsagainst GenBank and by detection of conserved proteindomains using RPS-BLAST [49] Tandem repeats withincontig sequences were identified using dotter [50] Theclassification of LTR retrotransposons into distinctlineages and clades was done using phylogenetic ana-lyses of their RT sequences [15] Alignment of RTsequences was carried out with ClustalX [51] and thephylogenetic trees were calculated using neighbour-join-ing method The trees were drawn and edited using theFigTree programMicrosatellite sequences were identified using Tandem
Repeats Finder [52] and TRAP [53] programs while aBioPerl script was used to identify ISBP loci [54] Identi-fication and classification of repetitive sequences withinBAC clones was done via PROFREP web server [45] uti-lizing repeat-specific databases of 454 reads prepared inthis study The server performs BLAST-based searchesagainst databases of whole-genome or repeat-specific454 reads and generates plots of similarity hits along thequery sequence (number of hits is proportional to copynumber of the query in the genome)
Preparation of probes for cytogenetic mappingPrimers specific for tandem repeats (Additional file 3)were designed from sequence contigs that carry tandem
Figure 4 Major groups of microsatellite DNA sequences identified in banana 454 reads
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 7 of 10
organized repetitive units Labeled probes were preparedby PCR on M acuminata rsquoCalcutta 4prime genomic DNAwith biotin- and digoxigenin-labeled nucleotides ThePCR premix contained 1times PCR buffer 1 mM MgCl2 02mM dNTPs 02 μM primers 05 U Taq polymerase(Finnzymes) and 10 - 15 ng template DNA PCR reac-tion was performed as follows initial denaturation of3 min at 94degC followed by 30 cycles of 1 min at 94degC50 s at 57degC and 50 s at 72degC and final extension step5 min at 72degCSpecific primers were also designed for reverse tran-
scriptase (RT) domains of different retroelements(Additional file 3) In the first step the RT domainswere amplified using PCR with a mix containing 1timesPCR buffer 15 mM MgCl2 02 mM dNTPs 02 μMprimers 05 U Taq polymerase (Finnzymes) and 10 -15 ng template DNA PCR products were checked bygel-electrophoresis cleaned up using paramagneticbeads Agencourt Ampure (Beckman Coulter) clonedinto TOPO vector (Invitrogen) and transformed intoelectro-competent E coli cells 48 recombinant clonesfor each retroelement were PCR amplified using M13primers and separated on the gel electrophoresisClones for each RT domain were than cleaned upusing ExoSAP-IT (USB Corporation) and used forSanger sequencing to verify presence of specific RTdomains in the clones The PCR products weresequenced with BigDye Terminator 31 Cycle Sequen-cing Kit (Applied Biosystems) on ABI 3730xl DNAanalyzer (Applied Biosystems) The nucleotidesequences were analyzed and edited using the StadenPackage [55] and searched for similarity with the cor-responding 454 contigs using BLAST [48] Clones withthe highest similarity to reconstructed contigs werePCR amplified with biotin- and digoxigenin-labelednucleotides and used as probes for fluorescence in situhybridization Selected clones used as probes showedat least 98 similarity with the corresponding 454sequence
Fluorescence in situ hybridization (FISH)FISH was done on mitotic metaphase spreads preparedfrom meristem root tip cells as described by Doleželovaacuteet al (1998) [56] The hybridization mixture consistedof 40 formamide 10 dextran sulfate in 1 times SSC anda 1 μgml labeled probe The mixture was added ontoslides and denatured at 80degC for 4 min The hybridiza-tion was carried out at 37degC overnight The sites ofprobe hybridization were detected using anti-digoxi-genin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories) and the chromosomes werecounterstained with DAPI The slides were examinedwith Olympus AX70 fluorescence microscope
Figure 5 Examples of repeat identification in M acuminataBAC clones Nucleotide sequences of BAC inserts are representedon X axis The plots represent genomic copy numbers of individualinsert regions calculated from numbers of similarity hits to 454 readdatabases The plot colors correspond to different types of therepeats (A) A lsquolow-copyrsquo BAC clone showing absence of repeatsalong its entire length (B) A BAC clone with relatively long stretchesof repetitive DNA (C) A highly repetitive BAC clone with varioustypes of repeats
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 8 of 10
(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

Ty3gypsy-like retrotransposons showed relatively lowdegree of phylogenetic diversity and most of thembelonged to the lineage of chromoviruses This singlelineage comprised about 87 of Ty3gypsy elementsidentified in this study thus greatly outnumbering ele-ments from the Tat lineage which included all otherTy3gypsy elements identified in the banana genomeThe chromoviral sequences could be classified into fourclades Galadriel Tekay Reina and CRM [30-32] Themost abundant chromoviral clade was Reina whichinvolved more than half of all chromoviral sequencesmaking up about 4 of the banana genome Many ele-ments belonging to this clade appeared to be non-autonomous as they lacked parts of RT-coding domain(data not shown) Members of the Tekay clade werefound to be the second most abundant group of chro-moviruses reaching about 2 of the genome Sequencesfrom the Galadriel clade corresponded to the retrotran-sposon monkey which has been identified earlier in thebanana genome [11] The consensus sequences of themonkey retrotransposon assembled from our 454 data asa 5880 bp fragment showed 95 similarity to the mon-key element described by Balint-Kurti et al (2000) [11]Previous estimates of the copy number using slot-blotanalysis indicated that monkey constituted about 02 -05 of the M acuminata genome [11] and are on linewith our estimates based on the proportion of monkey-derived sequences in 454 reads (Additional file 1)Although the monkey was supposed to be the mostabundant repetitive element in banana [5] our datashowed that several other families of retroelementsaccount for much larger parts of the genome The CRMclade sequences occupied a similar fraction of the gen-ome as those from the Galadriel clade Although beingmembers of the same evolutionary lineage banana chro-moviruses from distinct clades partly differed in their
chromosomal distribution Contrary to monkey whichpreferentially localized in secondary constrictions [11]members of other clades occupied mostly pericentro-meric regions and some additional loci in distal parts ofall chromosomes (Figures 3G H)
Non-LTR retrotransposons and DNA transposonsCompared to LTR-retrotransposons non-LTR retrotran-sposons and DNA transposons were found relativelyrare (Additional file 1) Within the clusters that repre-sented at least 001 of the genome only one cluster ofLINE sequences [33] and two clusters of DNA transpo-sons were identified The LINE elements were estimatedto constitute about 1 of the banana genome FISHwith a probe derived from reverse transcriptase domainof a LINE-like element resulted in dot signals in centro-meric regions on all chromosomes (Figure 3I) DNAtransposons identified in this work included elementsthat showed similarity to transposons belonging to thehAT superfamily [34] FISH with a probe derived fromhAT-related element failed to give visible signals mostprobably due to relatively small copy number The lowabundance of LINEs and DNA transposons seems to betypical for plant genomes and similar abundances wereobserved for example in rice grape and maize genomes[232435]
45S and 5S rDNAClusters containing 45S rDNA represented 112 of thegenome and the 45S rDNA sequence region was recon-structed as a 7553 bp fragment that included completesequence of the 18S-58S-26S rRNA locus surroundedby parts of intergenic spacer (IGS) Moreover based onsimilarity searches to BAC clone MA4_01C21 fromM acuminata which was sequenced within the contextof the Global Musa Genomics Consortium [13] and
Figure 1 Genome proportion of major groups of repetitive sequences identified in banana 454 data
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 3 of 10
which carries 45S rDNA units another cluster contain-ing IGS-like sequence was identified in the 454 dataIn contrast to Balint-Kurti et al (2000) whose results
obtained after FISH with mitotic chromosomes indi-cated insertion of a part of monkey into 45S rDNA [11]our 454 data suggests that monkey is not frequentlyassociated with the 18S-58S-26S rRNA gene copies
A plausible explanation for this discrepancy is that theinsertion is adjacent to the 45S rDNA locus In fact thespatial resolution of FISH on mitotic chromosomes isnot sufficient to discriminate two loci closer than 5 - 10Mbp [3637] A close vicinity of the monkey fragment to45S rDNA is supported by the sequence data of theBAC MA4_01C21 comprising 45S rDNA in which a
Figure 2 Phylogenetic analysis of Musa retrotransposons based on RT sequences Unrooted phylogenetic trees of Ty1copia elements (A)and Ty3gypsy elements (B) Names of the contigs assembled from 454 reads are printed in purple Classification of the Ty3gypsy lineages andchromoviral clades was done according to [30-32] Major lineages of Ty1copia elements were named according to a selected representative ofeach group
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 4 of 10
15kb fragment of monkey was identified However thefragment was not inserted in the 45SrDNA or IGSsequences The fact that this BAC comprises a chromo-virus element from the Tekay lineage and the SIRE1Maximus lineage indicates that the BAC MA4_01C21actually encompasses a border of the 45S rDNA locusand flanking genomic sequences characterized bysequence-heterogeneity and insertion of various mobileelementsSimilar to the 45S rRNA gene cluster our 454
sequence data enabled reconstruction of the entire cod-ing part of the 5S rRNA gene and its non-transcribedspacer The 5S rDNA was found to represent 038 ofthe banana nuclear genome Teo et al [38] identified aTy1copia-like element in the 5S rDNA spacer in severalbanana species The analysis of retrotransposon proteincoding domains in our data confirmed that the 5SrDNA spacer contained a part of the reverse transcrip-tase of the Tnt1-like element
Tandem organized repeatsRepeat reconstruction from the 454 data led to discov-ery of two new tandemly organized repeats One ofthem (CL33) consists of ~130 bp monomer while theCL18 repeat is characterized by ~2 kb monomer unitFISH on mitotic chromosomes revealed clusters of sig-nals in the subtelomeric regions of one pair of chromo-somes (satellite CL18) and weak signals in telomericregion on two pairs of chromosomes (satellite CL33)(Figures 3A B C) Southern hybridization resulted in aladder-like pattern typical for tandemly organized repeti-tive units for repeat CL33 only (not shown) The repeatCL18 gave a weak smear with a few visible bands mostlikely due to partially dispersed distribution andor poorconservation of the monomer length A rather low copynumber of CL33 andor long repetitive unit of satelliteCL18 may explain why these repeats were not identifiedin previous studies [56] In general the absence ofmore abundant tandem repeats in the banana genomemay be related to its relatively small size Satellite DNAis a typical component of subtelomeric and centromericchromosome regions in various plant species but theyoften form blocks of repeats in interstitial regions[1539-42] The results of this study as well as our ear-lier observations [56] indicate that typical centromericsatellite DNA is absent in the banana genome and thatthe centromeric regions are likely to be made of varioustypes of retrotransposons
Identification of DNA markersFollowing the thorough characterization of banana repe-titive DNA we screened the 454 sequences for the pre-sence of loci potentially suitable for use as DNAmarkers We focused on identification of simple
sequence repeats (SSRs) and sites of insertions of trans-posable elements (ISBP - Insertion Site Based Poly-morphism) [43] In total 27946 of 454 reads containingSSRs were identified with repeat units ranging from 2 to10 bp The most abundant motifs were dinucleotidesTA and GA and trinucleotides GAA (Figure 4) Morethan 11000 reads were identified to contain potentialISBP sites most of them carrying insertions of retro-transposons into unknown low-copy sequences Data-bases containing 454 reads carrying SSR sequences andpotential ISBPs were established and made publiclyavailable on our website [44]
Repeat identification in sequenced DNA clonesAs the next step in utilizing 454 data we took advantageof the read clustering during repeat analysis and createddatabases of sequence reads sorted according to theirrepeat of origin These databases were then utilized forsimilarity-based repeat detection and classification ingenomic BAC clone sequences implemented at thePROFREP server [45] The analysis was performed for49 BAC clones from M acuminata cv lsquoCalcutta 4prime andfor 12 BAC clones from M balbisiana cv lsquoPisang Klu-tug Wulungrsquo [14] The clones were sequenced as part ofthe Generation Challenge Program supported project(GCP-2005-15) and were selected based on the presenceof resistance gene homologs and other gene-likesequences Indeed out of the 49 M acuminata BACclones only 9 clones were highly repetitive 15 BACclones contained a single copy sequence with a largerepetitive region and the remaining BAC clones com-prised single copy sequence without any detectable repe-titive DNA andor carried a very short repetitive region(Figure 5) Out of the 12 BAC clones from M balbisi-ana two were single copy while the remaining 10 BACclones carried low copy sequences mixed with largerepetitive regions The repetitive profiles of all 62 BACclones are available as supplementary data (Additionalfile 2)
ConclusionsThis work represents a major advance in the analysis ofthe nuclear genome organization in banana an impor-tant staple and cash crop The application of low-depth454 sequencing provided until now the largest amountof DNA sequence data and enabled a detailed analysisof repetitive components of its nuclear genome Allmajor types of DNA repeats were characterized andMusa DNA repeats databases were established The ana-lysis of genomic distribution of selected repeats providednew data on long-range molecular organization ofbanana chromosomes and a large number of loci poten-tially useful as DNA markers were identified Theimproved knowledge and resources generated in this
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 5 of 10
Figure 3 Genomic distribution of different types of DNA repeats Mitotic metaphase spreads of M acuminata cv lsquoCalcutta 4rsquo (2n = 22) afterFISH with probes for various repeats The chromosomes were counterstained with DAPI (blue) Bar = 5 μm (A) Tandem repeat CL18 (greensignal) formed a cluster on one pair of chromosomes (long arrows) (B) Tandem repeat CL33 (red signal) localized on two pairs of chromosomes(long arrows) (C) Simultaneous hybridization of probes for CL18 (green signal) and CL33 (red signal long arrows) revealed co-localization of bothsatellites on one pair of chromosomes (short arrows) (D) Two metaphase plates after FISH with a probe for CL1SCL2Contig1080 - bananaretrotransposon belonging to SIREMaximus lineage (green) Uneven genomic distribution with clusters dispersed on all chromosomes isobvious (E) Similar genomic distribution was found for banana retroelement related to the Angela lineage (CL2Contig49) (F) Bananaretrotransposon belonging to Tnt1 lineage (CL10Contig16 red color) gave weak signals preferentially localized in distal parts of chromosomes(long arrows) (G) The most abundant type of Ty3gypsy-like element of the Reina lineage (CL1SCL5Contig891) localized preferentially tocentromeric or peri-centromeric regions of all chromosomes (green signals) (H) Also the Ty3gypsy-like element related to Tekay evolutionarylineage (CL4Contig82) clustered in centromeric or peri-centromeric regions of all chromosomes (I) A probe derived from LINE element(CL1SCL8Contig452) localized in the centromeric regions of all chromosomes (green signals)
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 6 of 10
study will be useful in annotating the banana genomesequence in the analysis of the evolution of the Musagenome and for the study of dynamics of DNA repeatsover evolutionary time scale as well as to isolate DNAmarkers for use in genetic diversity studies and in mar-ker-assisted selection
Methods454 sequencingIn vitro rooted plants of M acuminata cv lsquoCalcutta 4prime(ITC 0249) were obtained from the International TransitCentre (Bioversity International Global Musa Genebankhosted by the Katholieke Universiteit Leuven Belgium)and grown in a greenhouse DNA for sequencing wasprepared from nuclei isolated from healthy young leaftissues according to Zhang et al (1995) [46] Isolatednuclei were incubated with 40 mM EDTA 02 SDSand 025 μgμl proteinase K for 5 hours at 37degC andDNA was purified by phenolchloroform precipitationThe 454 sequencing was performed at the ArizonaGenomics Institute (Tucson USA) using 454 LifeSciencesRoche FLX instrument All sequence informa-tion generated in this study are available on our website[44] and was submitted to the National Center for Bio-technology Information short read archive under acces-sion numbers SRR057410 and SRR057411
Data analysisFollowing a removal of linkerprimer contaminationsand artificially duplicated reads the remaining 477699reads (average length of 206 nucleotides) were used forrepeat analysis The analysis was performed as describedby Macas et al (2007) [15] employing TGICL [47] anda set of custom-made BioPerl scripts for similarity-basedclustering and assembly of reads The clustering para-meters used by a tclust program (part of TGICL) were
set to consider pairwise similarity of two reads signifi-cant if it involved an overlap of at least 150 nucleotideswith 90 or better similarity representing at least 55and 70 of the length of longer and shorter read respec-tively (OVL = 150 PID = 90 LCOV = 55 SCOV = 70)The reads within individual clusters were assembledinto contigs using TGICL run with the -O lsquo-p 80 -o 40primeparameters specifying overlap percent identity andminimal length cutoff for cap3 assembler Repeat typeidentification was done using blastn and blastx [48]sequence-similarity searches of assembled contigsagainst GenBank and by detection of conserved proteindomains using RPS-BLAST [49] Tandem repeats withincontig sequences were identified using dotter [50] Theclassification of LTR retrotransposons into distinctlineages and clades was done using phylogenetic ana-lyses of their RT sequences [15] Alignment of RTsequences was carried out with ClustalX [51] and thephylogenetic trees were calculated using neighbour-join-ing method The trees were drawn and edited using theFigTree programMicrosatellite sequences were identified using Tandem
Repeats Finder [52] and TRAP [53] programs while aBioPerl script was used to identify ISBP loci [54] Identi-fication and classification of repetitive sequences withinBAC clones was done via PROFREP web server [45] uti-lizing repeat-specific databases of 454 reads prepared inthis study The server performs BLAST-based searchesagainst databases of whole-genome or repeat-specific454 reads and generates plots of similarity hits along thequery sequence (number of hits is proportional to copynumber of the query in the genome)
Preparation of probes for cytogenetic mappingPrimers specific for tandem repeats (Additional file 3)were designed from sequence contigs that carry tandem
Figure 4 Major groups of microsatellite DNA sequences identified in banana 454 reads
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 7 of 10
organized repetitive units Labeled probes were preparedby PCR on M acuminata rsquoCalcutta 4prime genomic DNAwith biotin- and digoxigenin-labeled nucleotides ThePCR premix contained 1times PCR buffer 1 mM MgCl2 02mM dNTPs 02 μM primers 05 U Taq polymerase(Finnzymes) and 10 - 15 ng template DNA PCR reac-tion was performed as follows initial denaturation of3 min at 94degC followed by 30 cycles of 1 min at 94degC50 s at 57degC and 50 s at 72degC and final extension step5 min at 72degCSpecific primers were also designed for reverse tran-
scriptase (RT) domains of different retroelements(Additional file 3) In the first step the RT domainswere amplified using PCR with a mix containing 1timesPCR buffer 15 mM MgCl2 02 mM dNTPs 02 μMprimers 05 U Taq polymerase (Finnzymes) and 10 -15 ng template DNA PCR products were checked bygel-electrophoresis cleaned up using paramagneticbeads Agencourt Ampure (Beckman Coulter) clonedinto TOPO vector (Invitrogen) and transformed intoelectro-competent E coli cells 48 recombinant clonesfor each retroelement were PCR amplified using M13primers and separated on the gel electrophoresisClones for each RT domain were than cleaned upusing ExoSAP-IT (USB Corporation) and used forSanger sequencing to verify presence of specific RTdomains in the clones The PCR products weresequenced with BigDye Terminator 31 Cycle Sequen-cing Kit (Applied Biosystems) on ABI 3730xl DNAanalyzer (Applied Biosystems) The nucleotidesequences were analyzed and edited using the StadenPackage [55] and searched for similarity with the cor-responding 454 contigs using BLAST [48] Clones withthe highest similarity to reconstructed contigs werePCR amplified with biotin- and digoxigenin-labelednucleotides and used as probes for fluorescence in situhybridization Selected clones used as probes showedat least 98 similarity with the corresponding 454sequence
Fluorescence in situ hybridization (FISH)FISH was done on mitotic metaphase spreads preparedfrom meristem root tip cells as described by Doleželovaacuteet al (1998) [56] The hybridization mixture consistedof 40 formamide 10 dextran sulfate in 1 times SSC anda 1 μgml labeled probe The mixture was added ontoslides and denatured at 80degC for 4 min The hybridiza-tion was carried out at 37degC overnight The sites ofprobe hybridization were detected using anti-digoxi-genin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories) and the chromosomes werecounterstained with DAPI The slides were examinedwith Olympus AX70 fluorescence microscope
Figure 5 Examples of repeat identification in M acuminataBAC clones Nucleotide sequences of BAC inserts are representedon X axis The plots represent genomic copy numbers of individualinsert regions calculated from numbers of similarity hits to 454 readdatabases The plot colors correspond to different types of therepeats (A) A lsquolow-copyrsquo BAC clone showing absence of repeatsalong its entire length (B) A BAC clone with relatively long stretchesof repetitive DNA (C) A highly repetitive BAC clone with varioustypes of repeats
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 8 of 10
(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

which carries 45S rDNA units another cluster contain-ing IGS-like sequence was identified in the 454 dataIn contrast to Balint-Kurti et al (2000) whose results
obtained after FISH with mitotic chromosomes indi-cated insertion of a part of monkey into 45S rDNA [11]our 454 data suggests that monkey is not frequentlyassociated with the 18S-58S-26S rRNA gene copies
A plausible explanation for this discrepancy is that theinsertion is adjacent to the 45S rDNA locus In fact thespatial resolution of FISH on mitotic chromosomes isnot sufficient to discriminate two loci closer than 5 - 10Mbp [3637] A close vicinity of the monkey fragment to45S rDNA is supported by the sequence data of theBAC MA4_01C21 comprising 45S rDNA in which a
Figure 2 Phylogenetic analysis of Musa retrotransposons based on RT sequences Unrooted phylogenetic trees of Ty1copia elements (A)and Ty3gypsy elements (B) Names of the contigs assembled from 454 reads are printed in purple Classification of the Ty3gypsy lineages andchromoviral clades was done according to [30-32] Major lineages of Ty1copia elements were named according to a selected representative ofeach group
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 4 of 10
15kb fragment of monkey was identified However thefragment was not inserted in the 45SrDNA or IGSsequences The fact that this BAC comprises a chromo-virus element from the Tekay lineage and the SIRE1Maximus lineage indicates that the BAC MA4_01C21actually encompasses a border of the 45S rDNA locusand flanking genomic sequences characterized bysequence-heterogeneity and insertion of various mobileelementsSimilar to the 45S rRNA gene cluster our 454
sequence data enabled reconstruction of the entire cod-ing part of the 5S rRNA gene and its non-transcribedspacer The 5S rDNA was found to represent 038 ofthe banana nuclear genome Teo et al [38] identified aTy1copia-like element in the 5S rDNA spacer in severalbanana species The analysis of retrotransposon proteincoding domains in our data confirmed that the 5SrDNA spacer contained a part of the reverse transcrip-tase of the Tnt1-like element
Tandem organized repeatsRepeat reconstruction from the 454 data led to discov-ery of two new tandemly organized repeats One ofthem (CL33) consists of ~130 bp monomer while theCL18 repeat is characterized by ~2 kb monomer unitFISH on mitotic chromosomes revealed clusters of sig-nals in the subtelomeric regions of one pair of chromo-somes (satellite CL18) and weak signals in telomericregion on two pairs of chromosomes (satellite CL33)(Figures 3A B C) Southern hybridization resulted in aladder-like pattern typical for tandemly organized repeti-tive units for repeat CL33 only (not shown) The repeatCL18 gave a weak smear with a few visible bands mostlikely due to partially dispersed distribution andor poorconservation of the monomer length A rather low copynumber of CL33 andor long repetitive unit of satelliteCL18 may explain why these repeats were not identifiedin previous studies [56] In general the absence ofmore abundant tandem repeats in the banana genomemay be related to its relatively small size Satellite DNAis a typical component of subtelomeric and centromericchromosome regions in various plant species but theyoften form blocks of repeats in interstitial regions[1539-42] The results of this study as well as our ear-lier observations [56] indicate that typical centromericsatellite DNA is absent in the banana genome and thatthe centromeric regions are likely to be made of varioustypes of retrotransposons
Identification of DNA markersFollowing the thorough characterization of banana repe-titive DNA we screened the 454 sequences for the pre-sence of loci potentially suitable for use as DNAmarkers We focused on identification of simple
sequence repeats (SSRs) and sites of insertions of trans-posable elements (ISBP - Insertion Site Based Poly-morphism) [43] In total 27946 of 454 reads containingSSRs were identified with repeat units ranging from 2 to10 bp The most abundant motifs were dinucleotidesTA and GA and trinucleotides GAA (Figure 4) Morethan 11000 reads were identified to contain potentialISBP sites most of them carrying insertions of retro-transposons into unknown low-copy sequences Data-bases containing 454 reads carrying SSR sequences andpotential ISBPs were established and made publiclyavailable on our website [44]
Repeat identification in sequenced DNA clonesAs the next step in utilizing 454 data we took advantageof the read clustering during repeat analysis and createddatabases of sequence reads sorted according to theirrepeat of origin These databases were then utilized forsimilarity-based repeat detection and classification ingenomic BAC clone sequences implemented at thePROFREP server [45] The analysis was performed for49 BAC clones from M acuminata cv lsquoCalcutta 4prime andfor 12 BAC clones from M balbisiana cv lsquoPisang Klu-tug Wulungrsquo [14] The clones were sequenced as part ofthe Generation Challenge Program supported project(GCP-2005-15) and were selected based on the presenceof resistance gene homologs and other gene-likesequences Indeed out of the 49 M acuminata BACclones only 9 clones were highly repetitive 15 BACclones contained a single copy sequence with a largerepetitive region and the remaining BAC clones com-prised single copy sequence without any detectable repe-titive DNA andor carried a very short repetitive region(Figure 5) Out of the 12 BAC clones from M balbisi-ana two were single copy while the remaining 10 BACclones carried low copy sequences mixed with largerepetitive regions The repetitive profiles of all 62 BACclones are available as supplementary data (Additionalfile 2)
ConclusionsThis work represents a major advance in the analysis ofthe nuclear genome organization in banana an impor-tant staple and cash crop The application of low-depth454 sequencing provided until now the largest amountof DNA sequence data and enabled a detailed analysisof repetitive components of its nuclear genome Allmajor types of DNA repeats were characterized andMusa DNA repeats databases were established The ana-lysis of genomic distribution of selected repeats providednew data on long-range molecular organization ofbanana chromosomes and a large number of loci poten-tially useful as DNA markers were identified Theimproved knowledge and resources generated in this
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 5 of 10
Figure 3 Genomic distribution of different types of DNA repeats Mitotic metaphase spreads of M acuminata cv lsquoCalcutta 4rsquo (2n = 22) afterFISH with probes for various repeats The chromosomes were counterstained with DAPI (blue) Bar = 5 μm (A) Tandem repeat CL18 (greensignal) formed a cluster on one pair of chromosomes (long arrows) (B) Tandem repeat CL33 (red signal) localized on two pairs of chromosomes(long arrows) (C) Simultaneous hybridization of probes for CL18 (green signal) and CL33 (red signal long arrows) revealed co-localization of bothsatellites on one pair of chromosomes (short arrows) (D) Two metaphase plates after FISH with a probe for CL1SCL2Contig1080 - bananaretrotransposon belonging to SIREMaximus lineage (green) Uneven genomic distribution with clusters dispersed on all chromosomes isobvious (E) Similar genomic distribution was found for banana retroelement related to the Angela lineage (CL2Contig49) (F) Bananaretrotransposon belonging to Tnt1 lineage (CL10Contig16 red color) gave weak signals preferentially localized in distal parts of chromosomes(long arrows) (G) The most abundant type of Ty3gypsy-like element of the Reina lineage (CL1SCL5Contig891) localized preferentially tocentromeric or peri-centromeric regions of all chromosomes (green signals) (H) Also the Ty3gypsy-like element related to Tekay evolutionarylineage (CL4Contig82) clustered in centromeric or peri-centromeric regions of all chromosomes (I) A probe derived from LINE element(CL1SCL8Contig452) localized in the centromeric regions of all chromosomes (green signals)
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 6 of 10
study will be useful in annotating the banana genomesequence in the analysis of the evolution of the Musagenome and for the study of dynamics of DNA repeatsover evolutionary time scale as well as to isolate DNAmarkers for use in genetic diversity studies and in mar-ker-assisted selection
Methods454 sequencingIn vitro rooted plants of M acuminata cv lsquoCalcutta 4prime(ITC 0249) were obtained from the International TransitCentre (Bioversity International Global Musa Genebankhosted by the Katholieke Universiteit Leuven Belgium)and grown in a greenhouse DNA for sequencing wasprepared from nuclei isolated from healthy young leaftissues according to Zhang et al (1995) [46] Isolatednuclei were incubated with 40 mM EDTA 02 SDSand 025 μgμl proteinase K for 5 hours at 37degC andDNA was purified by phenolchloroform precipitationThe 454 sequencing was performed at the ArizonaGenomics Institute (Tucson USA) using 454 LifeSciencesRoche FLX instrument All sequence informa-tion generated in this study are available on our website[44] and was submitted to the National Center for Bio-technology Information short read archive under acces-sion numbers SRR057410 and SRR057411
Data analysisFollowing a removal of linkerprimer contaminationsand artificially duplicated reads the remaining 477699reads (average length of 206 nucleotides) were used forrepeat analysis The analysis was performed as describedby Macas et al (2007) [15] employing TGICL [47] anda set of custom-made BioPerl scripts for similarity-basedclustering and assembly of reads The clustering para-meters used by a tclust program (part of TGICL) were
set to consider pairwise similarity of two reads signifi-cant if it involved an overlap of at least 150 nucleotideswith 90 or better similarity representing at least 55and 70 of the length of longer and shorter read respec-tively (OVL = 150 PID = 90 LCOV = 55 SCOV = 70)The reads within individual clusters were assembledinto contigs using TGICL run with the -O lsquo-p 80 -o 40primeparameters specifying overlap percent identity andminimal length cutoff for cap3 assembler Repeat typeidentification was done using blastn and blastx [48]sequence-similarity searches of assembled contigsagainst GenBank and by detection of conserved proteindomains using RPS-BLAST [49] Tandem repeats withincontig sequences were identified using dotter [50] Theclassification of LTR retrotransposons into distinctlineages and clades was done using phylogenetic ana-lyses of their RT sequences [15] Alignment of RTsequences was carried out with ClustalX [51] and thephylogenetic trees were calculated using neighbour-join-ing method The trees were drawn and edited using theFigTree programMicrosatellite sequences were identified using Tandem
Repeats Finder [52] and TRAP [53] programs while aBioPerl script was used to identify ISBP loci [54] Identi-fication and classification of repetitive sequences withinBAC clones was done via PROFREP web server [45] uti-lizing repeat-specific databases of 454 reads prepared inthis study The server performs BLAST-based searchesagainst databases of whole-genome or repeat-specific454 reads and generates plots of similarity hits along thequery sequence (number of hits is proportional to copynumber of the query in the genome)
Preparation of probes for cytogenetic mappingPrimers specific for tandem repeats (Additional file 3)were designed from sequence contigs that carry tandem
Figure 4 Major groups of microsatellite DNA sequences identified in banana 454 reads
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 7 of 10
organized repetitive units Labeled probes were preparedby PCR on M acuminata rsquoCalcutta 4prime genomic DNAwith biotin- and digoxigenin-labeled nucleotides ThePCR premix contained 1times PCR buffer 1 mM MgCl2 02mM dNTPs 02 μM primers 05 U Taq polymerase(Finnzymes) and 10 - 15 ng template DNA PCR reac-tion was performed as follows initial denaturation of3 min at 94degC followed by 30 cycles of 1 min at 94degC50 s at 57degC and 50 s at 72degC and final extension step5 min at 72degCSpecific primers were also designed for reverse tran-
scriptase (RT) domains of different retroelements(Additional file 3) In the first step the RT domainswere amplified using PCR with a mix containing 1timesPCR buffer 15 mM MgCl2 02 mM dNTPs 02 μMprimers 05 U Taq polymerase (Finnzymes) and 10 -15 ng template DNA PCR products were checked bygel-electrophoresis cleaned up using paramagneticbeads Agencourt Ampure (Beckman Coulter) clonedinto TOPO vector (Invitrogen) and transformed intoelectro-competent E coli cells 48 recombinant clonesfor each retroelement were PCR amplified using M13primers and separated on the gel electrophoresisClones for each RT domain were than cleaned upusing ExoSAP-IT (USB Corporation) and used forSanger sequencing to verify presence of specific RTdomains in the clones The PCR products weresequenced with BigDye Terminator 31 Cycle Sequen-cing Kit (Applied Biosystems) on ABI 3730xl DNAanalyzer (Applied Biosystems) The nucleotidesequences were analyzed and edited using the StadenPackage [55] and searched for similarity with the cor-responding 454 contigs using BLAST [48] Clones withthe highest similarity to reconstructed contigs werePCR amplified with biotin- and digoxigenin-labelednucleotides and used as probes for fluorescence in situhybridization Selected clones used as probes showedat least 98 similarity with the corresponding 454sequence
Fluorescence in situ hybridization (FISH)FISH was done on mitotic metaphase spreads preparedfrom meristem root tip cells as described by Doleželovaacuteet al (1998) [56] The hybridization mixture consistedof 40 formamide 10 dextran sulfate in 1 times SSC anda 1 μgml labeled probe The mixture was added ontoslides and denatured at 80degC for 4 min The hybridiza-tion was carried out at 37degC overnight The sites ofprobe hybridization were detected using anti-digoxi-genin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories) and the chromosomes werecounterstained with DAPI The slides were examinedwith Olympus AX70 fluorescence microscope
Figure 5 Examples of repeat identification in M acuminataBAC clones Nucleotide sequences of BAC inserts are representedon X axis The plots represent genomic copy numbers of individualinsert regions calculated from numbers of similarity hits to 454 readdatabases The plot colors correspond to different types of therepeats (A) A lsquolow-copyrsquo BAC clone showing absence of repeatsalong its entire length (B) A BAC clone with relatively long stretchesof repetitive DNA (C) A highly repetitive BAC clone with varioustypes of repeats
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 8 of 10
(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

15kb fragment of monkey was identified However thefragment was not inserted in the 45SrDNA or IGSsequences The fact that this BAC comprises a chromo-virus element from the Tekay lineage and the SIRE1Maximus lineage indicates that the BAC MA4_01C21actually encompasses a border of the 45S rDNA locusand flanking genomic sequences characterized bysequence-heterogeneity and insertion of various mobileelementsSimilar to the 45S rRNA gene cluster our 454
sequence data enabled reconstruction of the entire cod-ing part of the 5S rRNA gene and its non-transcribedspacer The 5S rDNA was found to represent 038 ofthe banana nuclear genome Teo et al [38] identified aTy1copia-like element in the 5S rDNA spacer in severalbanana species The analysis of retrotransposon proteincoding domains in our data confirmed that the 5SrDNA spacer contained a part of the reverse transcrip-tase of the Tnt1-like element
Tandem organized repeatsRepeat reconstruction from the 454 data led to discov-ery of two new tandemly organized repeats One ofthem (CL33) consists of ~130 bp monomer while theCL18 repeat is characterized by ~2 kb monomer unitFISH on mitotic chromosomes revealed clusters of sig-nals in the subtelomeric regions of one pair of chromo-somes (satellite CL18) and weak signals in telomericregion on two pairs of chromosomes (satellite CL33)(Figures 3A B C) Southern hybridization resulted in aladder-like pattern typical for tandemly organized repeti-tive units for repeat CL33 only (not shown) The repeatCL18 gave a weak smear with a few visible bands mostlikely due to partially dispersed distribution andor poorconservation of the monomer length A rather low copynumber of CL33 andor long repetitive unit of satelliteCL18 may explain why these repeats were not identifiedin previous studies [56] In general the absence ofmore abundant tandem repeats in the banana genomemay be related to its relatively small size Satellite DNAis a typical component of subtelomeric and centromericchromosome regions in various plant species but theyoften form blocks of repeats in interstitial regions[1539-42] The results of this study as well as our ear-lier observations [56] indicate that typical centromericsatellite DNA is absent in the banana genome and thatthe centromeric regions are likely to be made of varioustypes of retrotransposons
Identification of DNA markersFollowing the thorough characterization of banana repe-titive DNA we screened the 454 sequences for the pre-sence of loci potentially suitable for use as DNAmarkers We focused on identification of simple
sequence repeats (SSRs) and sites of insertions of trans-posable elements (ISBP - Insertion Site Based Poly-morphism) [43] In total 27946 of 454 reads containingSSRs were identified with repeat units ranging from 2 to10 bp The most abundant motifs were dinucleotidesTA and GA and trinucleotides GAA (Figure 4) Morethan 11000 reads were identified to contain potentialISBP sites most of them carrying insertions of retro-transposons into unknown low-copy sequences Data-bases containing 454 reads carrying SSR sequences andpotential ISBPs were established and made publiclyavailable on our website [44]
Repeat identification in sequenced DNA clonesAs the next step in utilizing 454 data we took advantageof the read clustering during repeat analysis and createddatabases of sequence reads sorted according to theirrepeat of origin These databases were then utilized forsimilarity-based repeat detection and classification ingenomic BAC clone sequences implemented at thePROFREP server [45] The analysis was performed for49 BAC clones from M acuminata cv lsquoCalcutta 4prime andfor 12 BAC clones from M balbisiana cv lsquoPisang Klu-tug Wulungrsquo [14] The clones were sequenced as part ofthe Generation Challenge Program supported project(GCP-2005-15) and were selected based on the presenceof resistance gene homologs and other gene-likesequences Indeed out of the 49 M acuminata BACclones only 9 clones were highly repetitive 15 BACclones contained a single copy sequence with a largerepetitive region and the remaining BAC clones com-prised single copy sequence without any detectable repe-titive DNA andor carried a very short repetitive region(Figure 5) Out of the 12 BAC clones from M balbisi-ana two were single copy while the remaining 10 BACclones carried low copy sequences mixed with largerepetitive regions The repetitive profiles of all 62 BACclones are available as supplementary data (Additionalfile 2)
ConclusionsThis work represents a major advance in the analysis ofthe nuclear genome organization in banana an impor-tant staple and cash crop The application of low-depth454 sequencing provided until now the largest amountof DNA sequence data and enabled a detailed analysisof repetitive components of its nuclear genome Allmajor types of DNA repeats were characterized andMusa DNA repeats databases were established The ana-lysis of genomic distribution of selected repeats providednew data on long-range molecular organization ofbanana chromosomes and a large number of loci poten-tially useful as DNA markers were identified Theimproved knowledge and resources generated in this
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 5 of 10
Figure 3 Genomic distribution of different types of DNA repeats Mitotic metaphase spreads of M acuminata cv lsquoCalcutta 4rsquo (2n = 22) afterFISH with probes for various repeats The chromosomes were counterstained with DAPI (blue) Bar = 5 μm (A) Tandem repeat CL18 (greensignal) formed a cluster on one pair of chromosomes (long arrows) (B) Tandem repeat CL33 (red signal) localized on two pairs of chromosomes(long arrows) (C) Simultaneous hybridization of probes for CL18 (green signal) and CL33 (red signal long arrows) revealed co-localization of bothsatellites on one pair of chromosomes (short arrows) (D) Two metaphase plates after FISH with a probe for CL1SCL2Contig1080 - bananaretrotransposon belonging to SIREMaximus lineage (green) Uneven genomic distribution with clusters dispersed on all chromosomes isobvious (E) Similar genomic distribution was found for banana retroelement related to the Angela lineage (CL2Contig49) (F) Bananaretrotransposon belonging to Tnt1 lineage (CL10Contig16 red color) gave weak signals preferentially localized in distal parts of chromosomes(long arrows) (G) The most abundant type of Ty3gypsy-like element of the Reina lineage (CL1SCL5Contig891) localized preferentially tocentromeric or peri-centromeric regions of all chromosomes (green signals) (H) Also the Ty3gypsy-like element related to Tekay evolutionarylineage (CL4Contig82) clustered in centromeric or peri-centromeric regions of all chromosomes (I) A probe derived from LINE element(CL1SCL8Contig452) localized in the centromeric regions of all chromosomes (green signals)
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 6 of 10
study will be useful in annotating the banana genomesequence in the analysis of the evolution of the Musagenome and for the study of dynamics of DNA repeatsover evolutionary time scale as well as to isolate DNAmarkers for use in genetic diversity studies and in mar-ker-assisted selection
Methods454 sequencingIn vitro rooted plants of M acuminata cv lsquoCalcutta 4prime(ITC 0249) were obtained from the International TransitCentre (Bioversity International Global Musa Genebankhosted by the Katholieke Universiteit Leuven Belgium)and grown in a greenhouse DNA for sequencing wasprepared from nuclei isolated from healthy young leaftissues according to Zhang et al (1995) [46] Isolatednuclei were incubated with 40 mM EDTA 02 SDSand 025 μgμl proteinase K for 5 hours at 37degC andDNA was purified by phenolchloroform precipitationThe 454 sequencing was performed at the ArizonaGenomics Institute (Tucson USA) using 454 LifeSciencesRoche FLX instrument All sequence informa-tion generated in this study are available on our website[44] and was submitted to the National Center for Bio-technology Information short read archive under acces-sion numbers SRR057410 and SRR057411
Data analysisFollowing a removal of linkerprimer contaminationsand artificially duplicated reads the remaining 477699reads (average length of 206 nucleotides) were used forrepeat analysis The analysis was performed as describedby Macas et al (2007) [15] employing TGICL [47] anda set of custom-made BioPerl scripts for similarity-basedclustering and assembly of reads The clustering para-meters used by a tclust program (part of TGICL) were
set to consider pairwise similarity of two reads signifi-cant if it involved an overlap of at least 150 nucleotideswith 90 or better similarity representing at least 55and 70 of the length of longer and shorter read respec-tively (OVL = 150 PID = 90 LCOV = 55 SCOV = 70)The reads within individual clusters were assembledinto contigs using TGICL run with the -O lsquo-p 80 -o 40primeparameters specifying overlap percent identity andminimal length cutoff for cap3 assembler Repeat typeidentification was done using blastn and blastx [48]sequence-similarity searches of assembled contigsagainst GenBank and by detection of conserved proteindomains using RPS-BLAST [49] Tandem repeats withincontig sequences were identified using dotter [50] Theclassification of LTR retrotransposons into distinctlineages and clades was done using phylogenetic ana-lyses of their RT sequences [15] Alignment of RTsequences was carried out with ClustalX [51] and thephylogenetic trees were calculated using neighbour-join-ing method The trees were drawn and edited using theFigTree programMicrosatellite sequences were identified using Tandem
Repeats Finder [52] and TRAP [53] programs while aBioPerl script was used to identify ISBP loci [54] Identi-fication and classification of repetitive sequences withinBAC clones was done via PROFREP web server [45] uti-lizing repeat-specific databases of 454 reads prepared inthis study The server performs BLAST-based searchesagainst databases of whole-genome or repeat-specific454 reads and generates plots of similarity hits along thequery sequence (number of hits is proportional to copynumber of the query in the genome)
Preparation of probes for cytogenetic mappingPrimers specific for tandem repeats (Additional file 3)were designed from sequence contigs that carry tandem
Figure 4 Major groups of microsatellite DNA sequences identified in banana 454 reads
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 7 of 10
organized repetitive units Labeled probes were preparedby PCR on M acuminata rsquoCalcutta 4prime genomic DNAwith biotin- and digoxigenin-labeled nucleotides ThePCR premix contained 1times PCR buffer 1 mM MgCl2 02mM dNTPs 02 μM primers 05 U Taq polymerase(Finnzymes) and 10 - 15 ng template DNA PCR reac-tion was performed as follows initial denaturation of3 min at 94degC followed by 30 cycles of 1 min at 94degC50 s at 57degC and 50 s at 72degC and final extension step5 min at 72degCSpecific primers were also designed for reverse tran-
scriptase (RT) domains of different retroelements(Additional file 3) In the first step the RT domainswere amplified using PCR with a mix containing 1timesPCR buffer 15 mM MgCl2 02 mM dNTPs 02 μMprimers 05 U Taq polymerase (Finnzymes) and 10 -15 ng template DNA PCR products were checked bygel-electrophoresis cleaned up using paramagneticbeads Agencourt Ampure (Beckman Coulter) clonedinto TOPO vector (Invitrogen) and transformed intoelectro-competent E coli cells 48 recombinant clonesfor each retroelement were PCR amplified using M13primers and separated on the gel electrophoresisClones for each RT domain were than cleaned upusing ExoSAP-IT (USB Corporation) and used forSanger sequencing to verify presence of specific RTdomains in the clones The PCR products weresequenced with BigDye Terminator 31 Cycle Sequen-cing Kit (Applied Biosystems) on ABI 3730xl DNAanalyzer (Applied Biosystems) The nucleotidesequences were analyzed and edited using the StadenPackage [55] and searched for similarity with the cor-responding 454 contigs using BLAST [48] Clones withthe highest similarity to reconstructed contigs werePCR amplified with biotin- and digoxigenin-labelednucleotides and used as probes for fluorescence in situhybridization Selected clones used as probes showedat least 98 similarity with the corresponding 454sequence
Fluorescence in situ hybridization (FISH)FISH was done on mitotic metaphase spreads preparedfrom meristem root tip cells as described by Doleželovaacuteet al (1998) [56] The hybridization mixture consistedof 40 formamide 10 dextran sulfate in 1 times SSC anda 1 μgml labeled probe The mixture was added ontoslides and denatured at 80degC for 4 min The hybridiza-tion was carried out at 37degC overnight The sites ofprobe hybridization were detected using anti-digoxi-genin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories) and the chromosomes werecounterstained with DAPI The slides were examinedwith Olympus AX70 fluorescence microscope
Figure 5 Examples of repeat identification in M acuminataBAC clones Nucleotide sequences of BAC inserts are representedon X axis The plots represent genomic copy numbers of individualinsert regions calculated from numbers of similarity hits to 454 readdatabases The plot colors correspond to different types of therepeats (A) A lsquolow-copyrsquo BAC clone showing absence of repeatsalong its entire length (B) A BAC clone with relatively long stretchesof repetitive DNA (C) A highly repetitive BAC clone with varioustypes of repeats
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 8 of 10
(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

Figure 3 Genomic distribution of different types of DNA repeats Mitotic metaphase spreads of M acuminata cv lsquoCalcutta 4rsquo (2n = 22) afterFISH with probes for various repeats The chromosomes were counterstained with DAPI (blue) Bar = 5 μm (A) Tandem repeat CL18 (greensignal) formed a cluster on one pair of chromosomes (long arrows) (B) Tandem repeat CL33 (red signal) localized on two pairs of chromosomes(long arrows) (C) Simultaneous hybridization of probes for CL18 (green signal) and CL33 (red signal long arrows) revealed co-localization of bothsatellites on one pair of chromosomes (short arrows) (D) Two metaphase plates after FISH with a probe for CL1SCL2Contig1080 - bananaretrotransposon belonging to SIREMaximus lineage (green) Uneven genomic distribution with clusters dispersed on all chromosomes isobvious (E) Similar genomic distribution was found for banana retroelement related to the Angela lineage (CL2Contig49) (F) Bananaretrotransposon belonging to Tnt1 lineage (CL10Contig16 red color) gave weak signals preferentially localized in distal parts of chromosomes(long arrows) (G) The most abundant type of Ty3gypsy-like element of the Reina lineage (CL1SCL5Contig891) localized preferentially tocentromeric or peri-centromeric regions of all chromosomes (green signals) (H) Also the Ty3gypsy-like element related to Tekay evolutionarylineage (CL4Contig82) clustered in centromeric or peri-centromeric regions of all chromosomes (I) A probe derived from LINE element(CL1SCL8Contig452) localized in the centromeric regions of all chromosomes (green signals)
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 6 of 10
study will be useful in annotating the banana genomesequence in the analysis of the evolution of the Musagenome and for the study of dynamics of DNA repeatsover evolutionary time scale as well as to isolate DNAmarkers for use in genetic diversity studies and in mar-ker-assisted selection
Methods454 sequencingIn vitro rooted plants of M acuminata cv lsquoCalcutta 4prime(ITC 0249) were obtained from the International TransitCentre (Bioversity International Global Musa Genebankhosted by the Katholieke Universiteit Leuven Belgium)and grown in a greenhouse DNA for sequencing wasprepared from nuclei isolated from healthy young leaftissues according to Zhang et al (1995) [46] Isolatednuclei were incubated with 40 mM EDTA 02 SDSand 025 μgμl proteinase K for 5 hours at 37degC andDNA was purified by phenolchloroform precipitationThe 454 sequencing was performed at the ArizonaGenomics Institute (Tucson USA) using 454 LifeSciencesRoche FLX instrument All sequence informa-tion generated in this study are available on our website[44] and was submitted to the National Center for Bio-technology Information short read archive under acces-sion numbers SRR057410 and SRR057411
Data analysisFollowing a removal of linkerprimer contaminationsand artificially duplicated reads the remaining 477699reads (average length of 206 nucleotides) were used forrepeat analysis The analysis was performed as describedby Macas et al (2007) [15] employing TGICL [47] anda set of custom-made BioPerl scripts for similarity-basedclustering and assembly of reads The clustering para-meters used by a tclust program (part of TGICL) were
set to consider pairwise similarity of two reads signifi-cant if it involved an overlap of at least 150 nucleotideswith 90 or better similarity representing at least 55and 70 of the length of longer and shorter read respec-tively (OVL = 150 PID = 90 LCOV = 55 SCOV = 70)The reads within individual clusters were assembledinto contigs using TGICL run with the -O lsquo-p 80 -o 40primeparameters specifying overlap percent identity andminimal length cutoff for cap3 assembler Repeat typeidentification was done using blastn and blastx [48]sequence-similarity searches of assembled contigsagainst GenBank and by detection of conserved proteindomains using RPS-BLAST [49] Tandem repeats withincontig sequences were identified using dotter [50] Theclassification of LTR retrotransposons into distinctlineages and clades was done using phylogenetic ana-lyses of their RT sequences [15] Alignment of RTsequences was carried out with ClustalX [51] and thephylogenetic trees were calculated using neighbour-join-ing method The trees were drawn and edited using theFigTree programMicrosatellite sequences were identified using Tandem
Repeats Finder [52] and TRAP [53] programs while aBioPerl script was used to identify ISBP loci [54] Identi-fication and classification of repetitive sequences withinBAC clones was done via PROFREP web server [45] uti-lizing repeat-specific databases of 454 reads prepared inthis study The server performs BLAST-based searchesagainst databases of whole-genome or repeat-specific454 reads and generates plots of similarity hits along thequery sequence (number of hits is proportional to copynumber of the query in the genome)
Preparation of probes for cytogenetic mappingPrimers specific for tandem repeats (Additional file 3)were designed from sequence contigs that carry tandem
Figure 4 Major groups of microsatellite DNA sequences identified in banana 454 reads
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 7 of 10
organized repetitive units Labeled probes were preparedby PCR on M acuminata rsquoCalcutta 4prime genomic DNAwith biotin- and digoxigenin-labeled nucleotides ThePCR premix contained 1times PCR buffer 1 mM MgCl2 02mM dNTPs 02 μM primers 05 U Taq polymerase(Finnzymes) and 10 - 15 ng template DNA PCR reac-tion was performed as follows initial denaturation of3 min at 94degC followed by 30 cycles of 1 min at 94degC50 s at 57degC and 50 s at 72degC and final extension step5 min at 72degCSpecific primers were also designed for reverse tran-
scriptase (RT) domains of different retroelements(Additional file 3) In the first step the RT domainswere amplified using PCR with a mix containing 1timesPCR buffer 15 mM MgCl2 02 mM dNTPs 02 μMprimers 05 U Taq polymerase (Finnzymes) and 10 -15 ng template DNA PCR products were checked bygel-electrophoresis cleaned up using paramagneticbeads Agencourt Ampure (Beckman Coulter) clonedinto TOPO vector (Invitrogen) and transformed intoelectro-competent E coli cells 48 recombinant clonesfor each retroelement were PCR amplified using M13primers and separated on the gel electrophoresisClones for each RT domain were than cleaned upusing ExoSAP-IT (USB Corporation) and used forSanger sequencing to verify presence of specific RTdomains in the clones The PCR products weresequenced with BigDye Terminator 31 Cycle Sequen-cing Kit (Applied Biosystems) on ABI 3730xl DNAanalyzer (Applied Biosystems) The nucleotidesequences were analyzed and edited using the StadenPackage [55] and searched for similarity with the cor-responding 454 contigs using BLAST [48] Clones withthe highest similarity to reconstructed contigs werePCR amplified with biotin- and digoxigenin-labelednucleotides and used as probes for fluorescence in situhybridization Selected clones used as probes showedat least 98 similarity with the corresponding 454sequence
Fluorescence in situ hybridization (FISH)FISH was done on mitotic metaphase spreads preparedfrom meristem root tip cells as described by Doleželovaacuteet al (1998) [56] The hybridization mixture consistedof 40 formamide 10 dextran sulfate in 1 times SSC anda 1 μgml labeled probe The mixture was added ontoslides and denatured at 80degC for 4 min The hybridiza-tion was carried out at 37degC overnight The sites ofprobe hybridization were detected using anti-digoxi-genin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories) and the chromosomes werecounterstained with DAPI The slides were examinedwith Olympus AX70 fluorescence microscope
Figure 5 Examples of repeat identification in M acuminataBAC clones Nucleotide sequences of BAC inserts are representedon X axis The plots represent genomic copy numbers of individualinsert regions calculated from numbers of similarity hits to 454 readdatabases The plot colors correspond to different types of therepeats (A) A lsquolow-copyrsquo BAC clone showing absence of repeatsalong its entire length (B) A BAC clone with relatively long stretchesof repetitive DNA (C) A highly repetitive BAC clone with varioustypes of repeats
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 8 of 10
(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

study will be useful in annotating the banana genomesequence in the analysis of the evolution of the Musagenome and for the study of dynamics of DNA repeatsover evolutionary time scale as well as to isolate DNAmarkers for use in genetic diversity studies and in mar-ker-assisted selection
Methods454 sequencingIn vitro rooted plants of M acuminata cv lsquoCalcutta 4prime(ITC 0249) were obtained from the International TransitCentre (Bioversity International Global Musa Genebankhosted by the Katholieke Universiteit Leuven Belgium)and grown in a greenhouse DNA for sequencing wasprepared from nuclei isolated from healthy young leaftissues according to Zhang et al (1995) [46] Isolatednuclei were incubated with 40 mM EDTA 02 SDSand 025 μgμl proteinase K for 5 hours at 37degC andDNA was purified by phenolchloroform precipitationThe 454 sequencing was performed at the ArizonaGenomics Institute (Tucson USA) using 454 LifeSciencesRoche FLX instrument All sequence informa-tion generated in this study are available on our website[44] and was submitted to the National Center for Bio-technology Information short read archive under acces-sion numbers SRR057410 and SRR057411
Data analysisFollowing a removal of linkerprimer contaminationsand artificially duplicated reads the remaining 477699reads (average length of 206 nucleotides) were used forrepeat analysis The analysis was performed as describedby Macas et al (2007) [15] employing TGICL [47] anda set of custom-made BioPerl scripts for similarity-basedclustering and assembly of reads The clustering para-meters used by a tclust program (part of TGICL) were
set to consider pairwise similarity of two reads signifi-cant if it involved an overlap of at least 150 nucleotideswith 90 or better similarity representing at least 55and 70 of the length of longer and shorter read respec-tively (OVL = 150 PID = 90 LCOV = 55 SCOV = 70)The reads within individual clusters were assembledinto contigs using TGICL run with the -O lsquo-p 80 -o 40primeparameters specifying overlap percent identity andminimal length cutoff for cap3 assembler Repeat typeidentification was done using blastn and blastx [48]sequence-similarity searches of assembled contigsagainst GenBank and by detection of conserved proteindomains using RPS-BLAST [49] Tandem repeats withincontig sequences were identified using dotter [50] Theclassification of LTR retrotransposons into distinctlineages and clades was done using phylogenetic ana-lyses of their RT sequences [15] Alignment of RTsequences was carried out with ClustalX [51] and thephylogenetic trees were calculated using neighbour-join-ing method The trees were drawn and edited using theFigTree programMicrosatellite sequences were identified using Tandem
Repeats Finder [52] and TRAP [53] programs while aBioPerl script was used to identify ISBP loci [54] Identi-fication and classification of repetitive sequences withinBAC clones was done via PROFREP web server [45] uti-lizing repeat-specific databases of 454 reads prepared inthis study The server performs BLAST-based searchesagainst databases of whole-genome or repeat-specific454 reads and generates plots of similarity hits along thequery sequence (number of hits is proportional to copynumber of the query in the genome)
Preparation of probes for cytogenetic mappingPrimers specific for tandem repeats (Additional file 3)were designed from sequence contigs that carry tandem
Figure 4 Major groups of microsatellite DNA sequences identified in banana 454 reads
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 7 of 10
organized repetitive units Labeled probes were preparedby PCR on M acuminata rsquoCalcutta 4prime genomic DNAwith biotin- and digoxigenin-labeled nucleotides ThePCR premix contained 1times PCR buffer 1 mM MgCl2 02mM dNTPs 02 μM primers 05 U Taq polymerase(Finnzymes) and 10 - 15 ng template DNA PCR reac-tion was performed as follows initial denaturation of3 min at 94degC followed by 30 cycles of 1 min at 94degC50 s at 57degC and 50 s at 72degC and final extension step5 min at 72degCSpecific primers were also designed for reverse tran-
scriptase (RT) domains of different retroelements(Additional file 3) In the first step the RT domainswere amplified using PCR with a mix containing 1timesPCR buffer 15 mM MgCl2 02 mM dNTPs 02 μMprimers 05 U Taq polymerase (Finnzymes) and 10 -15 ng template DNA PCR products were checked bygel-electrophoresis cleaned up using paramagneticbeads Agencourt Ampure (Beckman Coulter) clonedinto TOPO vector (Invitrogen) and transformed intoelectro-competent E coli cells 48 recombinant clonesfor each retroelement were PCR amplified using M13primers and separated on the gel electrophoresisClones for each RT domain were than cleaned upusing ExoSAP-IT (USB Corporation) and used forSanger sequencing to verify presence of specific RTdomains in the clones The PCR products weresequenced with BigDye Terminator 31 Cycle Sequen-cing Kit (Applied Biosystems) on ABI 3730xl DNAanalyzer (Applied Biosystems) The nucleotidesequences were analyzed and edited using the StadenPackage [55] and searched for similarity with the cor-responding 454 contigs using BLAST [48] Clones withthe highest similarity to reconstructed contigs werePCR amplified with biotin- and digoxigenin-labelednucleotides and used as probes for fluorescence in situhybridization Selected clones used as probes showedat least 98 similarity with the corresponding 454sequence
Fluorescence in situ hybridization (FISH)FISH was done on mitotic metaphase spreads preparedfrom meristem root tip cells as described by Doleželovaacuteet al (1998) [56] The hybridization mixture consistedof 40 formamide 10 dextran sulfate in 1 times SSC anda 1 μgml labeled probe The mixture was added ontoslides and denatured at 80degC for 4 min The hybridiza-tion was carried out at 37degC overnight The sites ofprobe hybridization were detected using anti-digoxi-genin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories) and the chromosomes werecounterstained with DAPI The slides were examinedwith Olympus AX70 fluorescence microscope
Figure 5 Examples of repeat identification in M acuminataBAC clones Nucleotide sequences of BAC inserts are representedon X axis The plots represent genomic copy numbers of individualinsert regions calculated from numbers of similarity hits to 454 readdatabases The plot colors correspond to different types of therepeats (A) A lsquolow-copyrsquo BAC clone showing absence of repeatsalong its entire length (B) A BAC clone with relatively long stretchesof repetitive DNA (C) A highly repetitive BAC clone with varioustypes of repeats
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 8 of 10
(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

organized repetitive units Labeled probes were preparedby PCR on M acuminata rsquoCalcutta 4prime genomic DNAwith biotin- and digoxigenin-labeled nucleotides ThePCR premix contained 1times PCR buffer 1 mM MgCl2 02mM dNTPs 02 μM primers 05 U Taq polymerase(Finnzymes) and 10 - 15 ng template DNA PCR reac-tion was performed as follows initial denaturation of3 min at 94degC followed by 30 cycles of 1 min at 94degC50 s at 57degC and 50 s at 72degC and final extension step5 min at 72degCSpecific primers were also designed for reverse tran-
scriptase (RT) domains of different retroelements(Additional file 3) In the first step the RT domainswere amplified using PCR with a mix containing 1timesPCR buffer 15 mM MgCl2 02 mM dNTPs 02 μMprimers 05 U Taq polymerase (Finnzymes) and 10 -15 ng template DNA PCR products were checked bygel-electrophoresis cleaned up using paramagneticbeads Agencourt Ampure (Beckman Coulter) clonedinto TOPO vector (Invitrogen) and transformed intoelectro-competent E coli cells 48 recombinant clonesfor each retroelement were PCR amplified using M13primers and separated on the gel electrophoresisClones for each RT domain were than cleaned upusing ExoSAP-IT (USB Corporation) and used forSanger sequencing to verify presence of specific RTdomains in the clones The PCR products weresequenced with BigDye Terminator 31 Cycle Sequen-cing Kit (Applied Biosystems) on ABI 3730xl DNAanalyzer (Applied Biosystems) The nucleotidesequences were analyzed and edited using the StadenPackage [55] and searched for similarity with the cor-responding 454 contigs using BLAST [48] Clones withthe highest similarity to reconstructed contigs werePCR amplified with biotin- and digoxigenin-labelednucleotides and used as probes for fluorescence in situhybridization Selected clones used as probes showedat least 98 similarity with the corresponding 454sequence
Fluorescence in situ hybridization (FISH)FISH was done on mitotic metaphase spreads preparedfrom meristem root tip cells as described by Doleželovaacuteet al (1998) [56] The hybridization mixture consistedof 40 formamide 10 dextran sulfate in 1 times SSC anda 1 μgml labeled probe The mixture was added ontoslides and denatured at 80degC for 4 min The hybridiza-tion was carried out at 37degC overnight The sites ofprobe hybridization were detected using anti-digoxi-genin-FITC (Roche Applied Science) and streptavidin-Cy3 (Vector Laboratories) and the chromosomes werecounterstained with DAPI The slides were examinedwith Olympus AX70 fluorescence microscope
Figure 5 Examples of repeat identification in M acuminataBAC clones Nucleotide sequences of BAC inserts are representedon X axis The plots represent genomic copy numbers of individualinsert regions calculated from numbers of similarity hits to 454 readdatabases The plot colors correspond to different types of therepeats (A) A lsquolow-copyrsquo BAC clone showing absence of repeatsalong its entire length (B) A BAC clone with relatively long stretchesof repetitive DNA (C) A highly repetitive BAC clone with varioustypes of repeats
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 8 of 10
(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

(Olympus) and the images of DAPI FITC and Cy-3fluorescence were acquired separately with a cooledhigh-resolution black and white CCD camera The cam-era was interfaced to a PC running the MicroImage soft-ware (Olympus)
Additional material
Additional file 1 Genome proportion of newly characterizedbanana repetitive elements Genome proportion of repetitive elementswas estimated from the sum of genome representation values (GR) ofcorresponding clusters of reads
Additional file 2 Repetitive profiles of sequenced BAC clones DNAsequence profiles of selected clones from three BAC libraries of Macuminata cv lsquoCalcutta 4rsquo (MA4 and C4BAM) M acuminata cvlsquoCavendishrsquo (MAC) and MBP BAC library from M balbisiana cv lsquoPisangKlutug Wulungrsquo httpolomoucuebcasczdna-librariesbananas
Additional file 3 Primers used for PCR amplification of satelliteDNA and different types of retrotransposons
AcknowledgementsWe thank Ir Ines van den Houwe for providing the plant material and MsRadka Tuškovaacute for excellent technical assistance This work was supported bythe Academy of Sciences of the Czech Republic (awards KJB500380901IAA600380703 and AVOZ50510513) the Czech Ministry of Education Youthand Sports (award LC06004) and the International Atomic Energy Agency(Research Agreement no 13192)
Author details1Laboratory of Molecular Cytogenetics and Cytometry Institute ofExperimental Botany Sokolovskaacute 6 Olomouc CZ-77200 Czech Republic2Biology Centre ASCR Institute of Plant Molecular Biology Branišovskaacute 31Českeacute Budĕjovice CZ-37005 Czech Republic 3National Institute ofAgrobiological Sciences Kannondai Tsukuba Ibaraki 305-8602 Japan4Commodities for Livelihoods Programme Bioversity International ParcScientifique Agropolis II 34397 Montpellier Cedex 5 France
Authorsrsquo contributionsEH isolated genomic DNA for sequencing and performed detailed repeatanalysis and their cytogenetic mapping JM performed initial 454 readclusteringassembly and PN conducted phylogenetic classification ofretrotransposon sequences TM sequenced BAC clones JD and JM made anintellectual contribution to the concept of the study and JD JM and NRrevised the manuscript critically for important intellectual content Allauthors read and approved the final manuscript
Received 8 December 2009 Accepted 16 September 2010Published 16 September 2010
References1 Food and Agriculture Organization (FAO) [httpfaostatfaoorg]2 Simonds NW Shepherd K The taxonomy and origins of the cultivated
bananas J Linn Soc Bot 1955 55302-3123 Doležel J Doleželovaacute M Novaacutek FJ Flow cytometric estimation of nuclear
DNA amount in diploid bananas (Musa acuminata and M balbisiana)Biol Plantarum 1994 36351-357
4 Bartoš J Alkhimova O Doleželovaacute M De Langhe E Doležel J Nucleargenome size and genomic distribution of ribosomal DNA in Musa andEnsete (Musaceae) taxonomic implications Cytogenet Genome Res 200510950-57
5 Hřibovaacute E Doleželovaacute M Town CD Macas J Doležel J Isolation andcharacterization of the highly repeated fraction of the banana genomeCytogenet Genome Res 2007 119268-274
6 Valaacuterik M Šimkovaacute H Hřibovaacute E Safaacuteř J Doleželovaacute M Doležel J Isolationcharacterization and chromosome localization of repetitive DNAsequences in bananas (Musa spp) Chromosome Res 2002 1089-100
7 Baurens FC Noyer JL Lanaud C Lagoda PJL A repetitive sequence familyof banana (Musa sp) shows homology to Copia-like elements J GenetBreed 1997 51135-142
8 Teo CH Tan SH Othman YR Schwarzacher T The cloning of Ty 1 -copia-like retrotransposons from 10 varieties of banana (Musa Sp) J BiochemMol Biol Biophys 2002 6193-201
9 Baurens FC Noyer JL Lanaud C Lagoda PJL Use of competitive PCR toessay copy number of repetitive elements in banana Mol Gen Genet1996 25357-64
10 Baurens FC Noyer JL Lanaud C Lagoda PJL Assessment of a species-specific element (Brep 1) in banana Theor Appl Genet 1997 95922-931
11 Balint-Kurti PJ Clendennen SK Doleželovaacute M Valaacuterik M Doležel JBeetham PR May GD Identification and chromosomal localization of themonkey retrotransposon in Musa sp Mol Gen Genet 2000 263908-915
12 Cheung F Town CD A BAC end view of the Musa acuminata genomeBMC Plant Biol 2007 729
13 Global Musa Genomics Consortium (GMGC) [httpwwwmusagenomicsorg]
14 Margulies M Egholm M Altman WE Attiya S Bader JS Bemben LA Berka JBraverman MS Chen YJ Chen ZT Dewell SB Du L Fierro JM Gomes XVGodwin BC He W Helgesen S Ho CH Irzyk GP Jando SC Alenquer MLIJarvie TP Jirage KB Kim JB Knight JR Lanza JR Leamon JH Lefkowitz SMLei M Li J Lohman KL Lu H Makhijani VB McDade KE McKenna MPMyers EW Nickerson E Nobile JR Plant R Puc BP Ronan MT Roth GTSarkis GJ Simons JF Simpson JW Srinivasan M Tartaro KR Tomasz AVogt KA Volkmer GA Wang SH Wang Y Weiner MP Yu PG Begley RFRothberg JM Genome sequencing in microfabricated high-densitypicolitre reactors Nature 2005 473376-380
15 Macas J Neumann P Navraacutetilovaacute A Repetitive DNA in the pea (Pisumsativum L) genome comprehensive characterization using 454sequencing and comparison to soybean and Medicago truncatula BMCGenomics 2007 8427
16 Swaminathan K Varala K Hudson ME Global repeat discovery andestimation of genomic copy number in a large complex genome usinga high-throughput 454 sequence survey BMC Genomics 2007 8132
17 Aert R Sagi L Volckaer G Gene content and density in banana (Musaacuminata) as revealed by genomic sequencing of BAC clones TheorAppl Genet 2004 109129-139
18 Azhar M Heslop-Harrison JS Genomes diversity and resistance geneanalogues in Musa species Cytogenet Genome Res 2008 12159-66
19 Vilarinhos AD Piffanelli P Lagoda P Thibivilliers S Sabau X Carreel FDHont A Construction and characterization of a bacterial artificialchromosome library of banana (Musa acuminata Colla) Theor Appl Genet2003 1061102-1106
20 Pillay M Ssebuliba R Hartman J Vuylsteke D Talengera DTushemereirwe W Conventional breeding strategies to enhance thesustainability of Musa biodiversity conservation for endemic cultivarsAfrican Crop Science Journal 2004 1259-65
21 Moens T Sandoval Fernandez JA Escalant JV De Waele D Evaluation ofthe progeny from a cross between lsquoPisang Berlinrsquo and M acuminata sppburmannicoides rsquoCalcutta 4prime for evidence of segregation with respect toresistance to black leaf streak disease and nematodes Infomusa 20021120-22
22 Bartoš J Paux E Kofler R Havraacutenkovaacute M Kopeckyacute D Suchaacutenkovaacute P Šafaacuteř JŠimkovaacute H Town CD Lelley T Feuillet C Doležel J A first survey of therye (Secale cereale) genome composition through BAC end sequencingof the short arm of chromosome 1R BMC Plant Biol 2008 895
23 International Rice Genome Sequencing Project The map-based sequenceof the rice genome Nature 2005 436793-800
24 Velasco R Zharkikh A Troggio M Cartwright DA Cestaro A Pruss DPindo M FitzGerald LM Vezzulli S Reid J Malacarne G Iliev D Coppola GWardell B Micheletti D Macalma T Facci M Mitchell JT Perazzolli MEldredge G Gatto P Oyzerski R Moretto M Gutin N Stefanini M Chen YSegala C Davenport C Dematte L Mraz A Battilana J Stormo K Costa FTao QZ Si-Ammour A Harkins T Lackey A Perbost C Taillon B Stella AFawcett JA Sterck L Vandepoele K Grando SM Toppo S Moser C
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 9 of 10
Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-

Lanchbury J Bogden R Skolnick M Sgaramella V Bhatnagar SK Fontana PGutin A Van de Peer Y Salamini F Viola R High quality draft consensussequence of the genome of a heterozygous grapevine variety PLoS ONE2007 2(12)e1326
25 Marin I Llorens C Ty3Gypsy retrotransposons Description of a newArabidopsis thaliana elements and evolutionary perspectives derivedfrom comparative genomics data Mol Biol Evol 2000 171040-1049
26 Havecker ER Gao X Voytas DF The Sireviruses a plant-specific lineage ofthe Ty1copia retrotransposons interact with a family of proteins relatedto dynein light chain 8 Plant Physiol 2005 139857-868
27 Wicker T Keller B Genome-wide comparative analysis of copiaretrotransposon in triticeae rice and Arabidopsis reveals conservedancient evolutionary lineages and distinct dynamics of individual copiafamilies Genome Res 2007 171072-1081
28 Grandbastien M-A Spielmann A Caboche M Tnt1 a mobile retroviral-liketransposable element of tobacco isolated by plant cell genetics Nature1989 337376-380
29 White SE Habera LF Wessler SR Retrotransposons in the flanking regionof normal plant genes A role for copia-like elements in the evolution ofgene structure and expression Proc Natl Acad Sci USA 19949111792-11796
30 Gorinsek B Gubensek F Kordis D Evolutionary genomics ofchromoviruses in eukaryotes Mol Biol Evol 2004 21781-798
31 Kordis D A genomic perspective on the chromodomain-containingretrotransposons Chromoviruses Gene 2005 347161-173
32 Llorens C Fares MA Moya A Relationships of gag-pol diversity betweenTy3Gypsy and Retroviridae LTR retroelements and the three kingshypothesis BMC Evol Biol 2008 8276
33 Schmidt T LINEs SINEs and repetitive DNA non-LTR retrotransposons inplant genomes Plant Mol Biol 1999 40903-910
34 Rubin E Lithwick G Levy AA Structure and evolution of the hATtransposon superfamily Genetics 2001 158949-957
35 Messing J Bharti AK Karlowski WM Gundlach H Kim HR Yu Y Wei FFuks G Soderlund CA Mayer KF Wing RA Sequence composition andgenome organization of maize Proc Natl Acad Sci USA 200410114349-14354
36 Raap AK Florijn RJ Blonden LJ Wiegant J Vaandrager JW Vrolijk H denDunnen J Tanke HJ Ommen GJ Fiber FISH as a DNA mapping toolMethods 1996 967-73
37 Pedersen C Linde-Laursen I The relationship between physical andgenetic distances at the Hor1 and Hor2 loci of barley estimated by two-colour fluorescent in situ hybridization Theor Appl Genet 1995 91941-946
38 Teo CH Schwarzacher T Tandem repeats and Musa chromosomeorganisation Unpublished GenBank code AM909712 - AM909714
39 Galasso I Schmidt T Pignone D Heslop-Harrison JS The molecularcytogenetics of Vigna unguiculata (L) Walp the physical organizationand characterization of 18s-58s-25s rRNA genes 5s rRNA genestelomere-like sequences and a family of centromeric repetitive DNAsequences Theor Appl Genet 1995 91928-935
40 Han YH Zhang ZH Liu JH Lu JY Huang SW Jin WW Distribution of thetandem repeat sequences and karyotyping in cucumber (Cucumissativus L) by fluorescence in situ hybridization Cytogenet Genome Res2008 12280-88
41 Jiang J Birchler JA Parrott WA Dawe RK A molecular view of plantcentromeres Trends Plant Sci 2003 8570-574
42 Nagaki K Tsujimoto H Sasakuma T A novel repetitive sequence of sugarcane SCEN family locating on centromeric regions Chromosome Res1998 6295-302
43 Paux E Roger D Badaeva E Gay G Bernard M Sourdille P Feuillet CCharacterizing the composition and evolution of homoeologousgenomes in hexaploid wheat through BAC-end sequencing onchromosome 3B Plant J 2006 48463-747
44 Laboratory of Molecular Cytogenetics and Cytomnetry (IEB CzechRepublic) [httpolomoucuebcasczbanana-sequencing-data]
45 Macas J Pech J Novaacutek P PROFREP a web server for repeat detection ingenomic sequences based on 454 sequencing data[httpw3lamcumbrcasczprofreppublic]
46 Zhang HB Zhao X Ding X Paterson AH Wing RA Preparation ofmegabase-size DNA from plant nuclei Plant J 1995 7175-184
47 Pertea G Huang X Liang F Antonescu V Sultana R Karamycheva S Lee YWhite J Cheung F Parvizi B Tsai J Quackenbush J TIGR Gene Indices
clustering tools (TGICL) a software system for fast clustering of largeEST datasets Bioinformatics 2003 19651-652
48 Altschul SF Madden TL Schaumlffer AA Zhang J Zhang Z Miller WLipman DJ Gapped BLAST and PSI-BLAST a new generation of proteindatabase search programs Nucleic Acids Res 1997 253389-3402
49 Marchler-Bauer A Anderson JB DeWeese-Scott C Fedorova ND Geer LYHe S Hurwitz DI Jackson JD Jacobs AR Lanczycki CJ Liebert CA Liu CMadej T Marchler GH Mazumder R Nikolskaya AN Panchenko AR Rao BSShoemaker BA Simonyan V Song JS Thiessen PA Vasudevan S Wang YYamashita RA Yin JJ Bryant SH CDD a curated Entrez database ofconserved domain alignments Nucleic Acids Res 2003 31383-387
50 Sonnhammer ELL Durbin R A dot-matrix program with dynamicthreshold control suited for genomic DNA and protein sequenceanalysis Gene 1995 167GC1-GC10
51 Thompson JD Gibson TJ Plewniak F Jeanmougin F Higgins DG TheCLUSTAL_X windows interface flexible strategies for multiple sequencealignment aided by quality analysis tools Nucleic Acids Res 1997254876-4882
52 Benson G Tandem repeats finder a program to analyze DNA sequencesNucleic Acids Res 1999 27573-580
53 Sobreira TJP Durham AM Gruber A TRAPautomated classificationquantification and annotation of tandemly repeated sequencesBioinformatics 2006 22361-362
54 Paux E Faure S Choulet F Roger D Gauthier V Martinant JP Sourdille PBalfourier F Le Paslier M-C Chauveau A Cakir M Gandon B Feuillet CInsertion site-based polymorphism markers open new perspectives forgenome saturation and marker-assisted selection in wheat PlantBiotechnol J 2010 8196-210
55 Staden R The Staden sequence analysis package Mol Biotechnol 19965233-241
56 Doleželovaacute M Valaacuterik M Swennen R Horry JP Doležel J Physical mappingof the 18S-25S and 5S ribosomal RNA genes in diploid bananas BiolPlantarum 1998 41497-505
doi1011861471-2229-10-204Cite this article as Hřibovaacute et al Repetitive part of the banana (Musaacuminata) genome investigated by low-depth 454 sequencing BMCPlant Biology 2010 10204
Submit your next manuscript to BioMed Centraland take full advantage of
bull Convenient online submission
bull Thorough peer review
bull No space constraints or color figure charges
bull Immediate publication on acceptance
bull Inclusion in PubMed CAS Scopus and Google Scholar
bull Research which is freely available for redistribution
Submit your manuscript at wwwbiomedcentralcomsubmit
Hřibovaacute et al BMC Plant Biology 2010 10204httpwwwbiomedcentralcom1471-222910204
Page 10 of 10
- Abstract
-
- Background
- Results
- Conclusion
-
- Background
- Results and Discussion
-
- LTR-retrotransposons
- Non-LTR retrotransposons and DNA transposons
- 45S and 5S rDNA
- Tandem organized repeats
- Identification of DNA markers
- Repeat identification in sequenced DNA clones
-
- Conclusions
- Methods
-
- 454 sequencing
- Data analysis
- Preparation of probes for cytogenetic mapping
- Fluorescence in situ hybridization (FISH)
-
- Acknowledgements
- Author details
- Authors contributions
- References
-