Research Article Concentration and Methylation of Cell-Free DNA...
Transcript of Research Article Concentration and Methylation of Cell-Free DNA...

Research ArticleConcentration and Methylation of Cell-Free DNA from BloodPlasma as Diagnostic Markers of Renal Cancer
Inessa Skrypkina1 Liudmyla Tsyba1 Kateryna Onyshchenko1
Dmytro Morderer1 Olena Kashparova1 Oleksii Nikolaienko1 Grigory Panasenko2
Sergii Vozianov3 Alina Romanenko3 and Alla Rynditch1
1Department of Functional Genomics Institute of Molecular Biology and Genetics of the National Academy of Science of UkraineKyiv Ukraine2Department of Molecular Oncogenetics Institute of Molecular Biology and Genetics of the National Academy of Science of UkraineKyiv Ukraine3Institute of Urology National Academy of Medical Sciences of Ukraine Kyiv Ukraine
Correspondence should be addressed to Inessa Skrypkina iskrypkinaimbgorgua
Received 10 June 2016 Revised 19 August 2016 Accepted 22 August 2016
Academic Editor Olav Lapaire
Copyright copy 2016 Inessa Skrypkina et al This is an open access article distributed under the Creative Commons AttributionLicense which permits unrestricted use distribution and reproduction in any medium provided the original work is properlycited
The critical point for successful treatment of cancer is diagnosis at early stages of tumor development Cancer cell-specificmethylated DNA has been found in the blood of cancer patients indicating that cell-free DNA (cfDNA) circulating in the blood isa convenient tumor-associated DNAmarker Therefore methylated cfDNA can be used as a minimally invasive diagnostic markerWe analysed the concentration of plasma cfDNA and methylation of six tumor suppressor genes in samples of 27 patients withrenal cancer and 15 healthy donors as controls The cfDNA concentrations in samples from cancer patients and healthy donors wasmeasured using two different methods the SYBR Green I fluorescence test and quantitative real-time PCR Both methods revealeda statistically significant increase of cfDNA concentrations in cancer patients Hypermethylation on cfDNA was detected for theLRRC3B (741) APC (519) FHIT (556) and RASSF1 (629) genes in patients with renal cancer Promoter methylation ofVHL and ITGA9 genes was not found on cfDNA Our results confirmed that the cfDNA level and methylation of CpG islands ofRASSF1A FHIT and APC genes in blood plasma can be used as noninvasive diagnostic markers of cancer
1 Introduction
Renal cell carcinoma (RCC) is awidespread oncologic diseasethat accounts for about 3 of all malignancies in adultsand 85 of all primarily malignant tumors in kidney [1]Metastases detected at the time of establishing a diagnosisare present in 25ndash30 of patients and even after surgerythe disease progresses in 20ndash30 of patients [2 3] Anasymptomatic period of the disease makes early diagnosisof this type of tumor difficult to perform Globally theincidence rates of kidney cancer are predicted to increaseTheInternational Agency for Research on Cancer claims that thisnumber will rise to 22 from 337860 cases in 2012 to 412929cases in 2020 [4]
Clear cell carcinoma is the most common type of RCCaccounting for 70ndash80 of all RCCs [5] Development ofthis particular type of RCC is associated with many tumorsuppressor genes that are localized in the short arm ofhuman chromosome 3 They can be inactivated as a resultof mutations LOH (loss of heterozygosity) or methylationof CpG islands in promoter regions [6ndash9] Identification ofaberrantly methylated genes for a particular tumor type canbe helpful in early diagnosis of the disease
Cell-free DNA (cfDNA) enters the blood stream fromapoptotic and necrotic tumor cells and is useful in detectingtumor-specific signatures including themethylation of genes[10 11] Aberrant cfDNA methylation has been described
Hindawi Publishing CorporationDisease MarkersVolume 2016 Article ID 3693096 10 pageshttpdxdoiorg10115520163693096
2 Disease Markers
in most cancer types and is being actively investigated forminimally invasive clinical diagnostics [11ndash13]
Large-scale NotI-microarray analyses of genetic and epi-genetic alterations in the genes of chromosome 3 in RCCrevealed that leucine-rich repeats containing 3B (LRRC3B)and Von Hippel-Lindau (VHL) genes possess the highest fre-quency of deletions andor methylations in renal carcinoma[14 15] Adenomatosis-polyposis-coli (APC) Ras associationdomain family 1 (RASSF1) and fragile histidine triad (FHIT)genes were shown to have high levels of methylation incfDNA andor in renal tumors [16ndash22]
In this study we determined the plasma cfDNA con-centration (by quantitative PCR and the fluorescence test)and analysed methylation of 6 genes (APC FHIT RASSF1LRRC3B VHL and ITGA9 (Integrin 12057291205731)) in plasma sam-ples from patients with kidney cancer in order to evaluate thediagnostic value of these markers for cancer detection
2 Materials and Methods
21 Sample Collection The study included 27 patients under-going surgery for kidney cancer at the Institute of UrologyNational Academy of Medical Sciences of Ukraine in Kyivbetween January 2011 and August 2011 Before surgery allpatients were fully examined according to the protocolsof the Ministry of Health of Ukraine laboratory clinicalanalysis Doppler ultrasound diagnosis renal scintigraphyand spiral computedmagnetic resonance tomography of theretroperitoneal space For the negative controls peripheralblood was collected from 15 healthy individuals All patientsgave written informed consent prior to enrollment in thestudy The samples were collected in accordance with theDeclaration of Helsinki and the guidelines issued by theEthics Committee of the Institute of Urology NAMS ofUkraine The Ethics Committee of the Institute of Urologyspecifically approved this study
22 Extraction of cfDNA Blood (5mL) was collected inK3 EDTA-containing tubes (Cat number 2102 APTACAItaly) The samples were stored at 4∘C and treated within 3 hafter blood collection The plasma was isolated by low-speedcentrifugation 250timesg for 7min 350timesg for 8min and 500timesgfor 10min using JouanMR23i centrifuge (JOUAN France) Itwas then aliquoted and cryopreserved at minus70∘C
cfDNA was isolated from 2mL plasma using the ProbaNA Kit (DNA-Technology Russia) according to the man-ufacturerrsquos recommendations (final elution volume was150 120583L) The extracted DNA was subjected to PCR with theACTB gene (51015840-CCACACTGTGCCCATCTACG-31015840 and 51015840-AGGATCTTCATGAGGTAGTCAGTCAG-31015840 99 bp frag-ment) as control and the PCR products were examinedby electrophoresis (see Supplementary Figure S1 in Sup-plementary Material available online at httpdxdoiorg10115520163693096) PCR conditions were as follows 95∘Cfor 4min and then 40 cycles of 95∘C for 40 s 56∘C for 20 sand 72∘C for 30 s with a final extension for 5min at 72∘C
23 Quantification of Plasma cfDNA by Real-Time PCRTo measure the plasma cfDNA concentration the genomic
sequence of 120573-actin was amplified by quantitative real-timePCR (qPCR) The primers and fluorescent probe used forqPCR were as described in Herrera et al [23] 5 120583L purifiedcfDNA was amplified using 03120583M of each primer (51015840-CCACACTGTGCCCATCTACG-31015840 and 51015840-AGGATCTTC-ATGAGGTAGTCAGTCAG-31015840) and a 025120583M fluorescentprobe (51015840-FAM-ATGCCCTCCCCCATGCCATCCTGCGT-TAMRA-31015840) The length of the amplified fragment was 99 bpPCR was performed under the following conditions 10minat 95∘C followed by 40 cycles of 15 s at 95∘C and 1minat 60∘C Quantitative standard curves were prepared usingserial dilutions (from 20 pg to 100 ngreaction) of controlgenomic DNA Human HCT116 DKO Nonmethylated DNA(Cat number D5014-1 Zymo Research Corporation USA)was used as calibrator for quantificationThe concentration ofcontrol DNA was assessed using a NanoDrop 2000 spectro-photometer (Thermo Scientific USA) No-template controls(NTCs) were used as negative controls The fluorescence ofthe amplified PCR products was detected using the BioRadiQ5 Optical System (Bio-Rad USA) The results of the qPCRassays represent the mean of three independent experi-ments each consisting of duplicate samples The analysiswas repeated if the difference between duplicate sampleswas greater than one cycle threshold The linear dynamicrange was determined by the standard curve and correlationcoefficients (1198772) which wasge098 Amore detailed version ofthe protocol is given in SupplementaryTable S1The statisticalsignificance of differences between samples was establishedusing the Mann-Whitney 119880 test
24 Quantification of Total Plasma DNA by the FluorescenceTest Evaluation of the cfDNA concentration was also per-formed measuring the fluorescence of intercalating dye [24]Specifically 5 120583L of a sample or the same volume of a standarddilution of genomicDNA (HumanHCT116DKONonmethy-lated DNA) with known concentration (0 ngmL and 9 serialdilutions from 1 to 256 ngmL) was added to 195 120583L of aSYBR Green I solution (Cat number S7585 Thermo FisherScientific USA) in PBS buffer (1 10000) and to black 96-well plates (PAA Cat number PAA30296X Austria) andincubated for 10min Two to three identical mixtures wereprepared from each sample or standard for greater accuracyThe fluorescence of the mixtures obtained was measured bythe ldquoVICTOR3 1420-050rdquo Multilabel Plate Readers (PerkinElmer USA) using filters for FITC (485535 nm) and 1 sacquisition time The DNA concentration was calculatedfrom the standard curve (1198772 was 097)
25 Evaluation of Gene Methylation Status Bisulfite treat-ment of isolated DNA was performed using the EZ DNAMethylation Kit (Cat number D5001 Zymo Research Cor-poration USA) according to the manufacturerrsquos instructionsThe methylation status of the different genes was determinedqualitatively by the methylation-specific polymerase chainreaction (MS-PCR) [25] Real-time MS-PCR was performedin a Bio-Rad iQ5 Real-Time PCR detection System (Bio-Rad USA) Primer sequences used for MS-PCR analysiswith PCR product size and primer annealing temperature
Disease Markers 3
are listed RASSF1 methylated-specific forward 51015840-GTG-TTAACGCGTTGCGTATC-31015840 and reverse 51015840-AACCCC-GCGAACTAAAAACGA-31015840 (60∘C 93 bp) [26] FHIT 51015840-TTGGGGCGCGGGTTTGGGTTTTTACGC-31015840 and 51015840-CGTAAACGACGCCGACCCCACTA-31015840 (62∘C 74 bp) [27]APC 51015840-TATTGCGGAGTGCGGGTC-31015840 and 51015840-TCGACG-AACTCCCGACGA-31015840 (60∘C 98 bp) [28] LRRC3B 51015840-GGTGCGAGGAAGGTAGGC-31015840 and 51015840-ACCAATACC-TCGCCGACG-31015840 (64∘C 149 bp) [29] VHL 51015840-TGGAGG-ATTTTTTTGCGTACGC-31015840 and 51015840-GAACCGAACGCC-GCGAA-31015840 (60∘C 158 bp) [30] ITGA9 51015840-TGGAGTATT-TTTACGATAATACGC-31015840 and 51015840-AAAAACCGAAAA-AACGACGA-31015840 (64∘C 116 bp) [31] Two 120583L of bisulfite-modified DNA was subjected to PCR amplification in a finalreaction volume of 25 120583L 1x Maxima SYBR Green qPCRMaster Mix (Cat number K0251 Thermo Scientific USA)and 03 120583M of each primer PCR was performed with aninitial 10min incubation at 95∘C followed by 45 cycles ofdenaturation at 95∘C for 15 s annealing for 20 s extensionat 72∘C for 30 s and a final 7min hold at 72∘C Each samplewas assayed in triplicate and each run included water blanksand an external control (universal methylated DNA) Afully methylated positive control was created by treating theDNA of lymphocytes from healthy donors with SssI CpGMethyltransferase (Cat number EM0821 Thermo ScientificUSA) according to themanufacturerrsquos recommendationsThespecificity of the PCR products was confirmed by meltingcurve analysis To verify MS-PCR data the MSP sequencingassay was performed using Genetic Analyser 3130 (AppliedBiosystems USA) following manufacturerrsquos protocols
26 Statistical Analysis Samples sizes were calculated usingthe formula described in [32] assuming 120572 and 120573 values of 005and 02 respectively We used standard deviation obtained inour preliminary experiments and estimated 150 differencein means
To evaluate the statistical significance of differencesbetween groups we performed the nonparametric Mann-Whitney 119880 test using the OriginPro 91 software (OriginLabUSA) or the Chi-square test (1205942) using Microsoft Excel 2007in the case of categorical variables
Differences were considered statistically significant if 119901 lt005 To evaluate the discriminative power of the parametersstudied for kidney cancer diagnostics we built binary logis-tic regression models for the selected predicting variablesand all their possible combinations using SPSS version 22(IBM USA) From these models the probabilities of positiveoutcome (ie cancer occurrence) were calculated Theseprobabilities were used for Receiver-operating characteristics(ROC) analysis Building of ROC and evaluation of AUC(Area Under Curve) was performed using the GraphPadPrism 607 (GraphPad Software La Jolla CA USA) or theOriginPro 91 software (OriginLab USA)
3 Results
31 Concentration of cfDNA in Blood Plasma of Patients withRenal Cancer and of Healthy Donors In this study bloodsamples from 27 patients with renal cancer and from 15
Table 1 Patient and tumor characteristics
Number of patientsAge at diagnosisAge gt 55 20 (741)Age lt 55 7 (259)
GenderMale 13 (481)Female 14 (519)
HistologyClear cell 23 (852)Sarcoma-like 2 (74)Papillary (75)clear cell (25) 1 (37)Cancer of the renal pelvis 1 (37)
Fuhrman gradeG1 + G2 19 (704)G3 + G4 8 (296)
Clinical stageStage 2 4 (148)Stage 3 23 (852)
Tumor-Node-Metastasis (TNM)T1a+b N0 M0-X 15 (556)T2 N0 M0-X 6 (222)T3 N0-1 M1-X 4 (148)TNM NA 2 (74)
healthy donors were used The blood samples were collectedbefore surgery in the Institute of Urology NAMS of UkraineThe results of the histological examination of tumors showedthat 23 patients had clear cell carcinoma 2 patients hadsarcoma-like tumors 1 patient had mixed type RCC (papil-laryclear cell) and 1 patient had cancer of the renal pelvis(Table 1)
The concentrations of cfDNA in blood plasma weredetermined by two methods by measuring the fluorescencelevel of intercalated SYBR Green I dye and by quantitativereal-time PCR (qPCR) of the 120573-actin gene
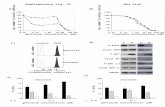
The results of the SYBR Green I fluorescence measure-ments showed that the concentrations of cfDNA in patientswith renal cancer range from 113 to 224912 ngmL (median23555 ngmL) The range of cfDNA concentration in healthydonors was much lower from 329 to 42675 ngmL (median5366 ngmL) (Figure 1(a))
qPCR revealed a statistically significant increaseof cfDNA concentration in cancer patients (median8097 ngmL range 233ndash11766 ngmL of plasma) As canbe seen from the box plot (Figure 1(c)) these values aresignificantly higher in RCC patients compared to healthydonors (median 351 ngmL 30ndash14678 ngmL of plasma)(Figure 1(c))
Receiver-operating characteristics (ROC) analysisshowed that the concentration of cfDNA can be used asdiagnostic feature for the detection of renal tumors (Figures1(b)ndash1(d)) AUC obtained for qRCR analysis was slightlyhigher (08049 119901 = 00012) than for the SYBR Green Ifluorescence measurements (07679 119901 = 00044) (Table 2)
4 Disease Markers
Table 2 Comparative analysis of different methods used to measure cfDNA in blood plasma
MethodqPCR analysis SYBR Green I fluorescence measurements
AUC 08049 (95 Cl 06602ndash09497) 07679 (95 Cl 06242ndash09116)Median (renal cancer) 8096 2355Median (control) 351 537119901 value (by Mann-Whitney 119880 test) 119901 lt 00008 119901 lt 00037
Renal cancer Control
0
1000
2000
Plas
ma D
NA
conc
entr
atio
n (n
gm
L)
p = 00037
(a)
0
50
100
Sens
itivi
ty (
)
50 1000100minus specificity ()
(b)
Renal cancer Control
p = 00008
0
500
1000
1500
Plas
ma D
NA
conc
entr
atio
n (n
gm
L)
(c)
50 10000
50
100
100minus specificity ()
Sens
itivi
ty (
)
(d)
Figure 1 Analysis of cfDNA concentration in plasma of patients with renal carcinoma and controls cfDNA concentrations were determinedby measuring the fluorescence level of intercalated SYBR Green I dye (a) and by qPCR (c) ROC curve analysis of cfDNA concentration incancer patients compared with the control group ((b) fluorescence test (d) qPCR)
32 Analysis of Methylation of Tumor Suppressor Genes incfDNA Since the cfDNA level alone cannot be a specificmarker of renal cancer [11] we also analysed the methylationstatus of CpG islands of 6 tumor suppressor genes in thecfDNA Using bisulfite treatment followed by MS-PCR wedetectedmethylation of theLRRC3BAPC FHIT andRASSF1genes in the cfDNA of cancer patients Promoter methylationof the LRRC3B gene was detected in 20 out of 27 samples(74) methylation of the RASSF1 APC and FHIT geneswas found in 17 (63) 14 (52) and 15 (556) patients
respectively (see Table 3 for detailed methylation frequen-cies)Methylation was not detected in the VHL and Integrin12057291205731 (ITGA9) genes in plasma cfDNA
Analysis of simultaneous methylation of CpG islands ofthe LRRC3B FHIT 0Pb and RASSF1 genes showed thatall the samples from cancer patients contained at least onemethylated promoter two promoters were methylated in333 three promoters were methylated in 27 and fourmethylated promoters were detected in 111 of the samples(Tables 3 and 4)
Disease Markers 5
Table 3 Summary of clinicopathological characteristics of patients with RCC and methylation status of LRRC3B RASSF1 FHIT and APCCpG islands in cfDNAlowast
Number Pathology Age (y) Sex pTNM Clinical grade Fuhrman nuclear grade MethylationLRRC3B RASSF1 APC FHIT
1 ccRCC 54 M af2N0M0 II 3 + + minus +2 ccRCC 61 M f1N0M0 II 2 + + minus +3 Sarcoma-like 66 F af2N0MX II 3 + minus + +4 PapillaryccRCC 63 M af1kN0MX III 1 minus + minus +5 ccRCC 47 F af31N0M1 III 3 + + + +6 ccRCC 64 M af31N0M1 III 3 + + + +7 ccRCC 58 M af2N0MX III 3 + minus + minus
8 ccRCC 61 M af1kN0M0 III 2 + + minus minus
9 ccRCC 75 M af1kN0MX III 2 minus + minus +10 ccRbb 65 M T2N0MX III 3 minus + + +11 ccRCC 61 F pT1N0M0 III 2 + minus + minus
12 ccRCC 63 F af31N1MX III 2-3 + minus + minus
13 ccRCC 68 F pT1 N0 MX III 1-2 + minus + +14 ccRCC 34 M af11N0 MX III 1 + minus + minus
15 Cancer of the renal pelvis 76 M pT3N0M1 III 4 + + + +16 ccRCC 56 F af11N0MX III 1 minus minus + +17 ccRCC 62 F af11N0MX III 1 + minus + +18 ccRCC 46 F af1kN0MX III 1 + + minus minus
19 ccRCC 55 F af2N0MX III 2 + + + minus
20 ccRCC 45 F T2N0M0 II 2 minus + minus minus
21 ccRCC 61 F af11N0MX III 2 + + minus minus
22 ccRCC 60 M NA III 2 + + minus +23 ccRCC 63 F af1kN0MX III 2 + minus minus minus
24 Sarcoma-like 60 F NA III 4 + minus minus minus
25 ccRCC 45 M pT1kN0MX III 2 minus + + +26 ccRCC 63 M af11N0M0 III 1 + + minus +27 ccRCC 73 F af11N0M0 III 1 minus + minus minus
lowastThe results in the Table are presented only for the genes with detected aberrant methylation in cfDNA
Table 4 Diagnostic data analysis for the discrimination of renal cancer patients and healthy subjects using cfDNA methylation of variousgenes alone and in combination
Markers Renal cell carcinoma (119899 = 27) Healthy controls (119899 = 15) 1205942 119901 value Sensitivitylowast Specificitylowastlowast LRRC3B 20 (741) 5 (333) 001 741 667RASSF1 17 (630 ) 1 (67) 00058 629 933FHIT 15 (556) 0 (0) 00003 556 100APC 14 (519) 1 (67) 00034 519 933VHL 0 (0) 0 (0) 0 100ITGA9 0 (0) 0 (0) 0 100RASSF1 or FHIT or APC 25 (923) 2 (133) lt00001 923 867RASSF1 or FHIT 21 (778) 1 (67) lt00001 778 933RASSF1 or APC 21 (778) 1 (67) lt00001 778 933lowastSensitivity was calculated as a percentage of positive results from a number of tested RCC patients lowastlowastspecificity was calculated as a percentage of negativetests from a given number of healthy donors
However LRRC3B showed a low specificity as a markerof cancer since it was methylated in 5 out of 15 (333)healthy donors Methylation of FHIT was not detected in thecfDNA of the control group while methylation of the APCand RASSF1 genes was found in 1 out of 15 (67) healthy
donors Methylation of APC and RASSF1 was detected indifferent healthy individuals (Table 4)
The sensitivity of each of thesemarkers exceeded 50 andwas 519 for APC 63 for RASSF1 and 556 for FHITwhich exhibited the best specificity in our test (100)
6 Disease Markers
Table 5 Receiver-operating characteristic (ROC) curve analyses of cfDNA marker-based models to discriminate between healthy persons(119899 = 15) and renal cancer patients (119899 = 27)lowast
AUC Std error 119901 value 95 LCL 95 UCLConc qPCR 080494 006771 000119 067223 093765Conc qPCR+APC 091852 004205 861E minus 06 08361 100094Conc qPCR+FHIT 091358 004898 110E minus 05 081759 100957Conc qPCR+RASSF1 088148 005986 500E minus 05 076416 099881Conc qPCR+ APC+RASSF1 1 0 106E minus 07 1 1Conc qPCR+APC+FHIT 095802 003018 112E minus 06 089888 101717Conc qPCR+RASSF1A+FHIT 094568 004521 216E minus 06 085708 103428Conc qPCR+APC+FHIT+RASSF1 1 0 106E minus 07 1 1lowastCalculated by binary logistic regression using combination of differentmarkers concentration of cfDNA determined by qPCR (Conc qPCR) andmethylationmarker genes (APC FHIT and RASSF1)
The use of the combined analysis of methylation statusof three genes (RASSF1 FHIT and APC) increased thesensitivity (778ndash923) while the specificity remained high(867ndash933) (Table 4) We did not find any correlationbetween hypermethylation cfDNA concentration and clin-icopathological parameters (grade lymph node metastasisage and sex) in patients with renal cancer
To explore the potential of combined cfDNA concen-tration and gene methylation for RCC diagnostics we per-formed binary logistic regression modelling As a predictorof variables we used cfDNA concentrations measured byquantitative PCR andmethylation ofAPC FHIT andRASSF1genes We built separate models for cfDNA concentrationalone and for all possible combinations of cfDNA concentra-tion and methylation of one two or three genes Predictiveproperties of the models were compared by ROC analysis Asreported above the AUC value for the cfDNA concentrationalone was 08 Addition of one of the genes slightly increasedthe AUC to values 088ndash0918 although these differenceswere not statistically significant as can be seen from 95confidence intervals (Table 5) Addition of two genes ledto further increase of the AUC value up to 1 when usingAPC and RASSF1 Finally the AUC value was 1 when weused the cfDNA concentration and methylation of all threegenes studied The results of ROC analysis are summarisedin Table 5 some of representative ROC curves are shown inFigure 2
4 Discussion
The level of cfDNA in blood plasma could be a universalmarker of malignancy [33] Many studies have shown thatchanges in cfDNA concentration can be correlated withdevelopment prognosis and survival of cancer patients Anincrease of cfDNA concentration was observed in patientswith breast gastric ovary lung colon and prostate cancer[11 34ndash39] It was suggested that an increase of cfDNAconcentration in cancer patients is associated with apoptosisand necrosis of cancer cells in the tumor microenvironment[40] This suggestion was supported by numerous cancer-specific alterations (such as allelic imbalances methylationand mutations) that were found in blood cfDNA (for reviews
00
02
04
06
08
10
Sens
itivi
ty
minus02 1000 02 06 0804
Conc_qPCR
Reference lineConc_qPCR APC+Conc_qPCR APC+ + FHIT
Conc_qPCR APC+ +FHIT + RASSF1A
1minus specificity
Figure 2 Receiver-operating characteristics (ROC) curves obtainedby using different models for the discrimination between healthycontrols (119899 = 15) and renal carcinoma patients (119899 = 27)Conc qPCR concentration of cfDNA determined by qPCR
see [11 41]) It was also demonstrated that monitoring of thecfDNA level in peripheral blood can be used as biomarker ofresponse to therapy in different cancer types [38 42 43]
Previous studies demonstrated that the evaluation ofconcentration of low molecular weight cfDNA (up to 100 bp)is the most representative for detection of malignancies anddisease prognosis since the level of fragments of this sizeincreases with disease progression [44ndash47] Recently Lu etal [47] showed that cfDNA fragments of 67 bp and 180 bpdid not differ between the controls and nonmetastatic RCCpatients while the cfDNA integrity index decreased fromcontrol to the metastatic group Significantly higher concen-trations of low molecular weight fragments were found inthe RCC patients [47] Here we have shown an increase ofcfDNA concentration in RCC patients using genomic cfDNAfragment of 120573-actin gene of 99 bp Recent experiments fromother laboratories also demonstrated increased cfDNA levels
Disease Markers 7
in the blood of patientswith renal cancer compared to healthyindividuals [19 48 49]
Absolute values of cfDNAconcentrations obtained by twodistinct methods are different (11ndash2249 ngmL of plasma inthe fluorescence test compared to 23ndash1177 ngmL in qPCR)but in both cases they were significantly higher than inhealthy individuals (4ndash426 ngmL in the fluorescence testand 3ndash146 ngmL in qPCR) The obtained results agree withdata from other studies in which the determination of thecfDNA concentration using fluorescent dyes gave highervalues than qPCR [24 41] The high AUC values obtainedfor both methods of cfDNA concentration measurement inRCC patients (07679 for fluorescent test and 080494 forqPCR) demonstrate that these methods can be used forclinical investigations In addition to quantitative changescfDNA also possesses qualitative changes that occur in DNAof tumor cells such as mutations microsatellite instabilityand methylation [11 41 50] Methylation of gene promotersis a well-known mechanism of regulation of gene expression[51] Most frequently aberrantmethylation of genes occurs incancer cells Aberrant methylation of the promoter detectedin cfDNA can be used for noninvasive detection of cancerdifferential diagnosis prognosis of survival and response tocancer therapy [52ndash57] Currently several diagnostic systemsbased on the detection of DNA methylation exist that areaimed not only at detecting malignancy (MethylMeter fromRiboMed USA) but also at detecting specific types of cancer(Epi proColon and Epi proLung from Epigenomics AG Ger-many Confirm MDx for Prostate Cancer from MDxHealthUSA Product series DecisionDx G-CIMP Melanom ECUMThymoma from Castle Biosciences) The search for newtools is being pursued because only two of these systems(MethylMeter and Epi proColon) can detect cancer at earlystages of development and can monitor treatment since theyare based on the detection of cfDNA methylation
In this study we started investigating methylation ofpreviously identified tumor suppressor genes in cfDNAData from many studies show that the RASSF1 gene playsan important role in cancerogenesis Hypermethylation ofRASSF1 CpG islands is associated with different types ofcancer and with the risk of progression of tumorigenesis [58ndash64] It was also shown that rat RASSF1 is involved in earlytumorigenesis of RCC [16 17] Studies on methylation of thisgene in blood serum led to controversial results Hauser et al[18] showed that RASSF1A is methylated in 229 of patientsthe study of De Martino et al [19] demonstrated methylationof RASSF1A in 459 of patients Hoque et al [20] observedmethylation of this gene in 11 of serum samples of patientswith RCC In our study methylation of RASSF1 was detectedin 629 of patients The differences in methylation levels ofthe RASSF1 gene can be explained by the use of different CpGislands for analyses We studied methylation of CpG regionlocated within the first exon of RASSF1C while Hauser et al[18] and De Martino et al [19] analysed the region locatedupstream of the initiation codon Previously it was reportedthat these two CpG islands were differentially methylatedin melanoma cell lines and melanoma tumors [65] Ellingeret al [66] demonstrated a 100 correlation between DNAhypermethylation of the RASSF1A promoter and papillary
RCC However De Martino et al [19] analysed 31 samplesof papillary RCCs and found no association of RASSF1Amethylation with the histological subtypes of RCC In ourstudy RASSF1 was also methylated in papillary RCC but itwas the only sample of this cancer subtype analysed
Previously we reported changes in the LRRC3B genepromoter during the search for genetic and epigenetic alter-ations in chromosome 3 in epithelial tumors using NotI-microarrays [14 67 68] LRRC3B was identified by Kim etal [69] as a putative gene suppressor of several tumors thatare silenced in gastric cancers by epigenetic mechanismsIncreased methylation of the LRRC3B gene promoter wasconfirmed in samples of clear cell RCC and colorectalhead and neck cancer [29 70 71] A high level of LRRC3Bhypermethylation was noted not only in RCC patients (74)but also in healthy donors (33) in our study questioning theuse of this gene for the diagnosis of renal cancer on cfDNA
The promoter of the APC gene was methylated in 519of patients which is in good agreement with the results ofHauser and colleagues [18] who detected methylation of theAPC gene in 543 of patients using cfDNA
Previously a significant correlation between FHITexpression in clear cell renal carcinomas and patientsurvival was demonstrated [21] Kvasha et al [22] showeda correlation between hypermethylation of the FHIT CpGisland and a significant decrease of FHIT expression inclear cell RCC The level of aberrant methylation of FHITobtained in our study on cfDNA (556) was close to theresults obtained in the study of Kvasha et al in samples ofRCC tumors (546)
Integrin 12057291205731 plays an important role in various signaltransduction pathways that control proliferation migrationand differentiation of both normal (reviewed in [72 73])and cancer cells (reviewed in [74 75]) Downregulation ofITGA9 expression was observed in several cancer types [76ndash78] that could be caused either by mutations in this gene [79]or by hypermethylation [31 68 80] However methylationof the ITGA9 gene was not detected in our experimentsWe also have not identified methylation of the VHL genealthough NotI-microarray hybridization revealed high levelsof changes in this gene (47) in renal cancer [14] It is possiblethat these changes are associated with deletions in the generather than with methylation At the same time in the studyof DeMartino et al where cfDNAwas analysed by restrictionanalysis methylated VHL was detected in 503 of patientswith RCC [19]
Methylation analysis of the RASSF1 FHIT and APCgenes demonstrated their high specificity (933 for RASSF1and APC 100 for FHIT) for renal tumors Neverthelesssensitivity in one gene analysis was just from 519 forAPC to 629 for RASSF1 (Table 4) At the same timethe use of a combination of three or two genes (withoutLRRC3B due to the low specificity of this gene) leads to asignificant increase in sensitivity (779ndash923) and specificity(867ndash933) All other combinations did not reveal anyadditional diagnosis information Simultaneous methylationof the RASSF1 APC and FHIT genes was identified only in3 patients with metastases However the small sample sizedoes not allow us to draw a conclusion on the correlation
8 Disease Markers
between methylation and disease progression At the sametime binary logistic regression analysis showed the consid-erable diagnostic potential of combining both approachesused in this study According to the ROC analysis the use ofonly cfDNA concentration has moderate diagnostic potential(AUC = 08) On the other hand by using the concentrationand methylation of two or three genes we achieved 100diagnostic accuracy in our samples These results of coursecannot be directly transferred to clinical practice and needverification on a larger number of samples However our datademonstrates the potential advantage there is in combiningevaluation of cfDNA concentration and gene methylation forRCC diagnostics and provides a basis for further research
Thus despite the small sampling our results confirm thepossibility of using the concentration of cfDNA in bloodplasma as an additional marker of renal cancer developmentand show that methylation of three genes FHIT APC andRASSF1 in cfDNA can be used to develop renal cancerdiagnostic tools
5 Conclusion
The results obtained indicate that the concentration of cell-free DNA in plasma and the methylation of specific genes(such as FHIT APC and RASSF1) can be a significantaddition to serological tumor markers in the identification ofpatients with renal cancer However further studies need tobe performed to evaluate their diagnostic value
Competing Interests
The authors declare that they have no competing interests
Acknowledgments
The authors thank Dr A-L Haenni for helpful discussionsand comments on the manuscript This work was supportedby Grants 0110U004534 and 115U002951 from NationalAcademy of Sciences of Ukraine
References
[1] G Banumathy and P Cairns ldquoSignaling pathways in renal cellcarcinomardquo Cancer Biology andTherapy vol 10 no 7 pp 658ndash664 2010
[2] K Gupta J D Miller J Z Li M W Russell and C Charbon-neau ldquoEpidemiologic and socioeconomic burden of metastaticrenal cell carcinoma (mRCC) a literature reviewrdquo CancerTreatment Reviews vol 34 no 3 pp 193ndash205 2008
[3] Ministry of Healthcare of Ukraine The Features Diagnosis andPrognosis Factors Renal Cell Carcinoma Ministry of Healthcareof Ukraine 2011 httpwwwsouuorgua
[4] World Cancer Research Fund InternationalAmerican Institutefor Cancer Research Continuous Update Project Report DietNutrition Physical Activity and Kidney Cancer 2015
[5] A Lopez-Beltran M Scarpelli R Montironi and Z Kirkalildquo2004 WHO classification of the renal tumors of the adultsrdquoEuropean Urology vol 49 no 5 pp 798ndash805 2006
[6] P Hadaczek Z Siprashvili M Markiewski et al ldquoAbsenceor reduction of FHIT expression in most clear cell renalcarcinomasrdquo Cancer Research vol 58 no 14 pp 2946ndash29511998
[7] E Arai and Y Kanai ldquoGenetic and epigenetic alterationsduring renal carcinogenesisrdquo International Journal of Clinicaland Experimental Pathology vol 4 no 1 pp 58ndash73 2011
[8] B N Lasseigne T C Burwell M A Patil D M Absher J DBrooks and R M Myers ldquoDNA methylation profiling revealsnovel diagnostic biomarkers in renal cell carcinomardquo BMCMedicine vol 12 no 1 article 235 2014
[9] N Shenoy N Vallumsetla Y Zou et al ldquoRole of DNAmethylation in renal cell carcinomardquo Journal of Hematology andOncology vol 8 no 1 article 88 2015
[10] G J Fournie J-P Courtin F Laval et al ldquoPlasma DNA asa marker of cancerous cell death Investigations in patientssuffering from lung cancer and in nude mice bearing humantumoursrdquo Cancer Letters vol 91 no 2 pp 221ndash227 1995
[11] H Schwarzenbach D S B Hoon and K Pantel ldquoCell-freenucleic acids as biomarkers in cancer patientsrdquo Nature ReviewsCancer vol 11 no 6 pp 426ndash437 2011
[12] B Taback and D S B Hoon ldquoCirculating nucleic acids andproteomics of plasmaserum clinical utilityrdquo Annals of the NewYork Academy of Sciences vol 1022 pp 1ndash8 2004
[13] R Wasserkort A Kalmar G Valcz et al ldquoAberrant septin 9DNA methylation in colorectal cancer is restricted to a singleCpG islandrdquo BMC Cancer vol 13 article 398 2013
[14] I Ya Skrypkina V I Kashuba V V Gordiyuk et al ldquoIdentifi-cation of changes in gene loci potentially associated with renalcancer by novel technique of NotI microarraysrdquo Reports of theNational Academy of Sciences of Ukraine vol 11 pp 188ndash1922006
[15] V I Kashuba I Y Skrypkina D V Saraev et al ldquoIdentificationof changes in gene loci potentially associated with cervical can-cer usingNotImicroarraysrdquoUkrainrsquoskyi Biokhimichnyi Zhurnalvol 78 no 2 pp 113ndash120 2006
[16] V I Loginov D S Khodyrev I V Pronina et al ldquoMethylationof the RASSF1A promoter region and the allelic imbalancefrequencies in chromosome 3 critical regions correlate withprogression of clear cell renal carcinomardquo Molecular Biologyvol 43 no 3 pp 394ndash402 2009
[17] I Peters K Rehmet N Wilke et al ldquoRASSF1A promotermethylation and expression analysis in normal and neoplastickidney indicates a role in early tumorigenesisrdquo MolecularCancer vol 6 article 49 2007
[18] S Hauser T Zahalka and G Fechner ldquoSerum DNA hyperme-thylation in patients with kidney cancer results of a prospectivestudyrdquo Anticancer Research vol 33 pp 4651ndash4656 2013
[19] M De Martino T Klatte A Haitel and M Marberger ldquoSerumcell-free DNA in renal cell carcinoma a diagnostic and prog-nostic markerrdquo Cancer vol 118 no 1 pp 82ndash90 2012
[20] M O Hoque S Begum O Topaloglu et al ldquoQuantitativedetection of promoter hypermethylation of multiple genes inthe tumor urine and serumDNAof patients with renal cancerrdquoCancer Research vol 64 no 15 pp 5511ndash5517 2004
[21] U Ramp E Caliskan T Ebert et al ldquoFHIT expression inclear cell renal carcinomas versatility of protein levels andcorrelation with survivalrdquoThe Journal of Pathology vol 196 no4 pp 430ndash436 2002
[22] S Kvasha V Gordiyuk A Kondratov et al ldquoHypermethylationof the 51015840CpG island of the FHIT gene in clear cell renalcarcinomasrdquo Cancer Letters vol 265 no 2 pp 250ndash257 2008
Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

2 Disease Markers
in most cancer types and is being actively investigated forminimally invasive clinical diagnostics [11ndash13]
Large-scale NotI-microarray analyses of genetic and epi-genetic alterations in the genes of chromosome 3 in RCCrevealed that leucine-rich repeats containing 3B (LRRC3B)and Von Hippel-Lindau (VHL) genes possess the highest fre-quency of deletions andor methylations in renal carcinoma[14 15] Adenomatosis-polyposis-coli (APC) Ras associationdomain family 1 (RASSF1) and fragile histidine triad (FHIT)genes were shown to have high levels of methylation incfDNA andor in renal tumors [16ndash22]
In this study we determined the plasma cfDNA con-centration (by quantitative PCR and the fluorescence test)and analysed methylation of 6 genes (APC FHIT RASSF1LRRC3B VHL and ITGA9 (Integrin 12057291205731)) in plasma sam-ples from patients with kidney cancer in order to evaluate thediagnostic value of these markers for cancer detection
2 Materials and Methods
21 Sample Collection The study included 27 patients under-going surgery for kidney cancer at the Institute of UrologyNational Academy of Medical Sciences of Ukraine in Kyivbetween January 2011 and August 2011 Before surgery allpatients were fully examined according to the protocolsof the Ministry of Health of Ukraine laboratory clinicalanalysis Doppler ultrasound diagnosis renal scintigraphyand spiral computedmagnetic resonance tomography of theretroperitoneal space For the negative controls peripheralblood was collected from 15 healthy individuals All patientsgave written informed consent prior to enrollment in thestudy The samples were collected in accordance with theDeclaration of Helsinki and the guidelines issued by theEthics Committee of the Institute of Urology NAMS ofUkraine The Ethics Committee of the Institute of Urologyspecifically approved this study
22 Extraction of cfDNA Blood (5mL) was collected inK3 EDTA-containing tubes (Cat number 2102 APTACAItaly) The samples were stored at 4∘C and treated within 3 hafter blood collection The plasma was isolated by low-speedcentrifugation 250timesg for 7min 350timesg for 8min and 500timesgfor 10min using JouanMR23i centrifuge (JOUAN France) Itwas then aliquoted and cryopreserved at minus70∘C
cfDNA was isolated from 2mL plasma using the ProbaNA Kit (DNA-Technology Russia) according to the man-ufacturerrsquos recommendations (final elution volume was150 120583L) The extracted DNA was subjected to PCR with theACTB gene (51015840-CCACACTGTGCCCATCTACG-31015840 and 51015840-AGGATCTTCATGAGGTAGTCAGTCAG-31015840 99 bp frag-ment) as control and the PCR products were examinedby electrophoresis (see Supplementary Figure S1 in Sup-plementary Material available online at httpdxdoiorg10115520163693096) PCR conditions were as follows 95∘Cfor 4min and then 40 cycles of 95∘C for 40 s 56∘C for 20 sand 72∘C for 30 s with a final extension for 5min at 72∘C
23 Quantification of Plasma cfDNA by Real-Time PCRTo measure the plasma cfDNA concentration the genomic
sequence of 120573-actin was amplified by quantitative real-timePCR (qPCR) The primers and fluorescent probe used forqPCR were as described in Herrera et al [23] 5 120583L purifiedcfDNA was amplified using 03120583M of each primer (51015840-CCACACTGTGCCCATCTACG-31015840 and 51015840-AGGATCTTC-ATGAGGTAGTCAGTCAG-31015840) and a 025120583M fluorescentprobe (51015840-FAM-ATGCCCTCCCCCATGCCATCCTGCGT-TAMRA-31015840) The length of the amplified fragment was 99 bpPCR was performed under the following conditions 10minat 95∘C followed by 40 cycles of 15 s at 95∘C and 1minat 60∘C Quantitative standard curves were prepared usingserial dilutions (from 20 pg to 100 ngreaction) of controlgenomic DNA Human HCT116 DKO Nonmethylated DNA(Cat number D5014-1 Zymo Research Corporation USA)was used as calibrator for quantificationThe concentration ofcontrol DNA was assessed using a NanoDrop 2000 spectro-photometer (Thermo Scientific USA) No-template controls(NTCs) were used as negative controls The fluorescence ofthe amplified PCR products was detected using the BioRadiQ5 Optical System (Bio-Rad USA) The results of the qPCRassays represent the mean of three independent experi-ments each consisting of duplicate samples The analysiswas repeated if the difference between duplicate sampleswas greater than one cycle threshold The linear dynamicrange was determined by the standard curve and correlationcoefficients (1198772) which wasge098 Amore detailed version ofthe protocol is given in SupplementaryTable S1The statisticalsignificance of differences between samples was establishedusing the Mann-Whitney 119880 test
24 Quantification of Total Plasma DNA by the FluorescenceTest Evaluation of the cfDNA concentration was also per-formed measuring the fluorescence of intercalating dye [24]Specifically 5 120583L of a sample or the same volume of a standarddilution of genomicDNA (HumanHCT116DKONonmethy-lated DNA) with known concentration (0 ngmL and 9 serialdilutions from 1 to 256 ngmL) was added to 195 120583L of aSYBR Green I solution (Cat number S7585 Thermo FisherScientific USA) in PBS buffer (1 10000) and to black 96-well plates (PAA Cat number PAA30296X Austria) andincubated for 10min Two to three identical mixtures wereprepared from each sample or standard for greater accuracyThe fluorescence of the mixtures obtained was measured bythe ldquoVICTOR3 1420-050rdquo Multilabel Plate Readers (PerkinElmer USA) using filters for FITC (485535 nm) and 1 sacquisition time The DNA concentration was calculatedfrom the standard curve (1198772 was 097)
25 Evaluation of Gene Methylation Status Bisulfite treat-ment of isolated DNA was performed using the EZ DNAMethylation Kit (Cat number D5001 Zymo Research Cor-poration USA) according to the manufacturerrsquos instructionsThe methylation status of the different genes was determinedqualitatively by the methylation-specific polymerase chainreaction (MS-PCR) [25] Real-time MS-PCR was performedin a Bio-Rad iQ5 Real-Time PCR detection System (Bio-Rad USA) Primer sequences used for MS-PCR analysiswith PCR product size and primer annealing temperature
Disease Markers 3
are listed RASSF1 methylated-specific forward 51015840-GTG-TTAACGCGTTGCGTATC-31015840 and reverse 51015840-AACCCC-GCGAACTAAAAACGA-31015840 (60∘C 93 bp) [26] FHIT 51015840-TTGGGGCGCGGGTTTGGGTTTTTACGC-31015840 and 51015840-CGTAAACGACGCCGACCCCACTA-31015840 (62∘C 74 bp) [27]APC 51015840-TATTGCGGAGTGCGGGTC-31015840 and 51015840-TCGACG-AACTCCCGACGA-31015840 (60∘C 98 bp) [28] LRRC3B 51015840-GGTGCGAGGAAGGTAGGC-31015840 and 51015840-ACCAATACC-TCGCCGACG-31015840 (64∘C 149 bp) [29] VHL 51015840-TGGAGG-ATTTTTTTGCGTACGC-31015840 and 51015840-GAACCGAACGCC-GCGAA-31015840 (60∘C 158 bp) [30] ITGA9 51015840-TGGAGTATT-TTTACGATAATACGC-31015840 and 51015840-AAAAACCGAAAA-AACGACGA-31015840 (64∘C 116 bp) [31] Two 120583L of bisulfite-modified DNA was subjected to PCR amplification in a finalreaction volume of 25 120583L 1x Maxima SYBR Green qPCRMaster Mix (Cat number K0251 Thermo Scientific USA)and 03 120583M of each primer PCR was performed with aninitial 10min incubation at 95∘C followed by 45 cycles ofdenaturation at 95∘C for 15 s annealing for 20 s extensionat 72∘C for 30 s and a final 7min hold at 72∘C Each samplewas assayed in triplicate and each run included water blanksand an external control (universal methylated DNA) Afully methylated positive control was created by treating theDNA of lymphocytes from healthy donors with SssI CpGMethyltransferase (Cat number EM0821 Thermo ScientificUSA) according to themanufacturerrsquos recommendationsThespecificity of the PCR products was confirmed by meltingcurve analysis To verify MS-PCR data the MSP sequencingassay was performed using Genetic Analyser 3130 (AppliedBiosystems USA) following manufacturerrsquos protocols
26 Statistical Analysis Samples sizes were calculated usingthe formula described in [32] assuming 120572 and 120573 values of 005and 02 respectively We used standard deviation obtained inour preliminary experiments and estimated 150 differencein means
To evaluate the statistical significance of differencesbetween groups we performed the nonparametric Mann-Whitney 119880 test using the OriginPro 91 software (OriginLabUSA) or the Chi-square test (1205942) using Microsoft Excel 2007in the case of categorical variables
Differences were considered statistically significant if 119901 lt005 To evaluate the discriminative power of the parametersstudied for kidney cancer diagnostics we built binary logis-tic regression models for the selected predicting variablesand all their possible combinations using SPSS version 22(IBM USA) From these models the probabilities of positiveoutcome (ie cancer occurrence) were calculated Theseprobabilities were used for Receiver-operating characteristics(ROC) analysis Building of ROC and evaluation of AUC(Area Under Curve) was performed using the GraphPadPrism 607 (GraphPad Software La Jolla CA USA) or theOriginPro 91 software (OriginLab USA)
3 Results
31 Concentration of cfDNA in Blood Plasma of Patients withRenal Cancer and of Healthy Donors In this study bloodsamples from 27 patients with renal cancer and from 15
Table 1 Patient and tumor characteristics
Number of patientsAge at diagnosisAge gt 55 20 (741)Age lt 55 7 (259)
GenderMale 13 (481)Female 14 (519)
HistologyClear cell 23 (852)Sarcoma-like 2 (74)Papillary (75)clear cell (25) 1 (37)Cancer of the renal pelvis 1 (37)
Fuhrman gradeG1 + G2 19 (704)G3 + G4 8 (296)
Clinical stageStage 2 4 (148)Stage 3 23 (852)
Tumor-Node-Metastasis (TNM)T1a+b N0 M0-X 15 (556)T2 N0 M0-X 6 (222)T3 N0-1 M1-X 4 (148)TNM NA 2 (74)
healthy donors were used The blood samples were collectedbefore surgery in the Institute of Urology NAMS of UkraineThe results of the histological examination of tumors showedthat 23 patients had clear cell carcinoma 2 patients hadsarcoma-like tumors 1 patient had mixed type RCC (papil-laryclear cell) and 1 patient had cancer of the renal pelvis(Table 1)
The concentrations of cfDNA in blood plasma weredetermined by two methods by measuring the fluorescencelevel of intercalated SYBR Green I dye and by quantitativereal-time PCR (qPCR) of the 120573-actin gene
The results of the SYBR Green I fluorescence measure-ments showed that the concentrations of cfDNA in patientswith renal cancer range from 113 to 224912 ngmL (median23555 ngmL) The range of cfDNA concentration in healthydonors was much lower from 329 to 42675 ngmL (median5366 ngmL) (Figure 1(a))
qPCR revealed a statistically significant increaseof cfDNA concentration in cancer patients (median8097 ngmL range 233ndash11766 ngmL of plasma) As canbe seen from the box plot (Figure 1(c)) these values aresignificantly higher in RCC patients compared to healthydonors (median 351 ngmL 30ndash14678 ngmL of plasma)(Figure 1(c))
Receiver-operating characteristics (ROC) analysisshowed that the concentration of cfDNA can be used asdiagnostic feature for the detection of renal tumors (Figures1(b)ndash1(d)) AUC obtained for qRCR analysis was slightlyhigher (08049 119901 = 00012) than for the SYBR Green Ifluorescence measurements (07679 119901 = 00044) (Table 2)
4 Disease Markers
Table 2 Comparative analysis of different methods used to measure cfDNA in blood plasma
MethodqPCR analysis SYBR Green I fluorescence measurements
AUC 08049 (95 Cl 06602ndash09497) 07679 (95 Cl 06242ndash09116)Median (renal cancer) 8096 2355Median (control) 351 537119901 value (by Mann-Whitney 119880 test) 119901 lt 00008 119901 lt 00037
Renal cancer Control
0
1000
2000
Plas
ma D
NA
conc
entr
atio
n (n
gm
L)
p = 00037
(a)
0
50
100
Sens
itivi
ty (
)
50 1000100minus specificity ()
(b)
Renal cancer Control
p = 00008
0
500
1000
1500
Plas
ma D
NA
conc
entr
atio
n (n
gm
L)
(c)
50 10000
50
100
100minus specificity ()
Sens
itivi
ty (
)
(d)
Figure 1 Analysis of cfDNA concentration in plasma of patients with renal carcinoma and controls cfDNA concentrations were determinedby measuring the fluorescence level of intercalated SYBR Green I dye (a) and by qPCR (c) ROC curve analysis of cfDNA concentration incancer patients compared with the control group ((b) fluorescence test (d) qPCR)
32 Analysis of Methylation of Tumor Suppressor Genes incfDNA Since the cfDNA level alone cannot be a specificmarker of renal cancer [11] we also analysed the methylationstatus of CpG islands of 6 tumor suppressor genes in thecfDNA Using bisulfite treatment followed by MS-PCR wedetectedmethylation of theLRRC3BAPC FHIT andRASSF1genes in the cfDNA of cancer patients Promoter methylationof the LRRC3B gene was detected in 20 out of 27 samples(74) methylation of the RASSF1 APC and FHIT geneswas found in 17 (63) 14 (52) and 15 (556) patients
respectively (see Table 3 for detailed methylation frequen-cies)Methylation was not detected in the VHL and Integrin12057291205731 (ITGA9) genes in plasma cfDNA
Analysis of simultaneous methylation of CpG islands ofthe LRRC3B FHIT 0Pb and RASSF1 genes showed thatall the samples from cancer patients contained at least onemethylated promoter two promoters were methylated in333 three promoters were methylated in 27 and fourmethylated promoters were detected in 111 of the samples(Tables 3 and 4)
Disease Markers 5
Table 3 Summary of clinicopathological characteristics of patients with RCC and methylation status of LRRC3B RASSF1 FHIT and APCCpG islands in cfDNAlowast
Number Pathology Age (y) Sex pTNM Clinical grade Fuhrman nuclear grade MethylationLRRC3B RASSF1 APC FHIT
1 ccRCC 54 M af2N0M0 II 3 + + minus +2 ccRCC 61 M f1N0M0 II 2 + + minus +3 Sarcoma-like 66 F af2N0MX II 3 + minus + +4 PapillaryccRCC 63 M af1kN0MX III 1 minus + minus +5 ccRCC 47 F af31N0M1 III 3 + + + +6 ccRCC 64 M af31N0M1 III 3 + + + +7 ccRCC 58 M af2N0MX III 3 + minus + minus
8 ccRCC 61 M af1kN0M0 III 2 + + minus minus
9 ccRCC 75 M af1kN0MX III 2 minus + minus +10 ccRbb 65 M T2N0MX III 3 minus + + +11 ccRCC 61 F pT1N0M0 III 2 + minus + minus
12 ccRCC 63 F af31N1MX III 2-3 + minus + minus
13 ccRCC 68 F pT1 N0 MX III 1-2 + minus + +14 ccRCC 34 M af11N0 MX III 1 + minus + minus
15 Cancer of the renal pelvis 76 M pT3N0M1 III 4 + + + +16 ccRCC 56 F af11N0MX III 1 minus minus + +17 ccRCC 62 F af11N0MX III 1 + minus + +18 ccRCC 46 F af1kN0MX III 1 + + minus minus
19 ccRCC 55 F af2N0MX III 2 + + + minus
20 ccRCC 45 F T2N0M0 II 2 minus + minus minus
21 ccRCC 61 F af11N0MX III 2 + + minus minus
22 ccRCC 60 M NA III 2 + + minus +23 ccRCC 63 F af1kN0MX III 2 + minus minus minus
24 Sarcoma-like 60 F NA III 4 + minus minus minus
25 ccRCC 45 M pT1kN0MX III 2 minus + + +26 ccRCC 63 M af11N0M0 III 1 + + minus +27 ccRCC 73 F af11N0M0 III 1 minus + minus minus
lowastThe results in the Table are presented only for the genes with detected aberrant methylation in cfDNA
Table 4 Diagnostic data analysis for the discrimination of renal cancer patients and healthy subjects using cfDNA methylation of variousgenes alone and in combination
Markers Renal cell carcinoma (119899 = 27) Healthy controls (119899 = 15) 1205942 119901 value Sensitivitylowast Specificitylowastlowast LRRC3B 20 (741) 5 (333) 001 741 667RASSF1 17 (630 ) 1 (67) 00058 629 933FHIT 15 (556) 0 (0) 00003 556 100APC 14 (519) 1 (67) 00034 519 933VHL 0 (0) 0 (0) 0 100ITGA9 0 (0) 0 (0) 0 100RASSF1 or FHIT or APC 25 (923) 2 (133) lt00001 923 867RASSF1 or FHIT 21 (778) 1 (67) lt00001 778 933RASSF1 or APC 21 (778) 1 (67) lt00001 778 933lowastSensitivity was calculated as a percentage of positive results from a number of tested RCC patients lowastlowastspecificity was calculated as a percentage of negativetests from a given number of healthy donors
However LRRC3B showed a low specificity as a markerof cancer since it was methylated in 5 out of 15 (333)healthy donors Methylation of FHIT was not detected in thecfDNA of the control group while methylation of the APCand RASSF1 genes was found in 1 out of 15 (67) healthy
donors Methylation of APC and RASSF1 was detected indifferent healthy individuals (Table 4)
The sensitivity of each of thesemarkers exceeded 50 andwas 519 for APC 63 for RASSF1 and 556 for FHITwhich exhibited the best specificity in our test (100)
6 Disease Markers
Table 5 Receiver-operating characteristic (ROC) curve analyses of cfDNA marker-based models to discriminate between healthy persons(119899 = 15) and renal cancer patients (119899 = 27)lowast
AUC Std error 119901 value 95 LCL 95 UCLConc qPCR 080494 006771 000119 067223 093765Conc qPCR+APC 091852 004205 861E minus 06 08361 100094Conc qPCR+FHIT 091358 004898 110E minus 05 081759 100957Conc qPCR+RASSF1 088148 005986 500E minus 05 076416 099881Conc qPCR+ APC+RASSF1 1 0 106E minus 07 1 1Conc qPCR+APC+FHIT 095802 003018 112E minus 06 089888 101717Conc qPCR+RASSF1A+FHIT 094568 004521 216E minus 06 085708 103428Conc qPCR+APC+FHIT+RASSF1 1 0 106E minus 07 1 1lowastCalculated by binary logistic regression using combination of differentmarkers concentration of cfDNA determined by qPCR (Conc qPCR) andmethylationmarker genes (APC FHIT and RASSF1)
The use of the combined analysis of methylation statusof three genes (RASSF1 FHIT and APC) increased thesensitivity (778ndash923) while the specificity remained high(867ndash933) (Table 4) We did not find any correlationbetween hypermethylation cfDNA concentration and clin-icopathological parameters (grade lymph node metastasisage and sex) in patients with renal cancer
To explore the potential of combined cfDNA concen-tration and gene methylation for RCC diagnostics we per-formed binary logistic regression modelling As a predictorof variables we used cfDNA concentrations measured byquantitative PCR andmethylation ofAPC FHIT andRASSF1genes We built separate models for cfDNA concentrationalone and for all possible combinations of cfDNA concentra-tion and methylation of one two or three genes Predictiveproperties of the models were compared by ROC analysis Asreported above the AUC value for the cfDNA concentrationalone was 08 Addition of one of the genes slightly increasedthe AUC to values 088ndash0918 although these differenceswere not statistically significant as can be seen from 95confidence intervals (Table 5) Addition of two genes ledto further increase of the AUC value up to 1 when usingAPC and RASSF1 Finally the AUC value was 1 when weused the cfDNA concentration and methylation of all threegenes studied The results of ROC analysis are summarisedin Table 5 some of representative ROC curves are shown inFigure 2
4 Discussion
The level of cfDNA in blood plasma could be a universalmarker of malignancy [33] Many studies have shown thatchanges in cfDNA concentration can be correlated withdevelopment prognosis and survival of cancer patients Anincrease of cfDNA concentration was observed in patientswith breast gastric ovary lung colon and prostate cancer[11 34ndash39] It was suggested that an increase of cfDNAconcentration in cancer patients is associated with apoptosisand necrosis of cancer cells in the tumor microenvironment[40] This suggestion was supported by numerous cancer-specific alterations (such as allelic imbalances methylationand mutations) that were found in blood cfDNA (for reviews
00
02
04
06
08
10
Sens
itivi
ty
minus02 1000 02 06 0804
Conc_qPCR
Reference lineConc_qPCR APC+Conc_qPCR APC+ + FHIT
Conc_qPCR APC+ +FHIT + RASSF1A
1minus specificity
Figure 2 Receiver-operating characteristics (ROC) curves obtainedby using different models for the discrimination between healthycontrols (119899 = 15) and renal carcinoma patients (119899 = 27)Conc qPCR concentration of cfDNA determined by qPCR
see [11 41]) It was also demonstrated that monitoring of thecfDNA level in peripheral blood can be used as biomarker ofresponse to therapy in different cancer types [38 42 43]
Previous studies demonstrated that the evaluation ofconcentration of low molecular weight cfDNA (up to 100 bp)is the most representative for detection of malignancies anddisease prognosis since the level of fragments of this sizeincreases with disease progression [44ndash47] Recently Lu etal [47] showed that cfDNA fragments of 67 bp and 180 bpdid not differ between the controls and nonmetastatic RCCpatients while the cfDNA integrity index decreased fromcontrol to the metastatic group Significantly higher concen-trations of low molecular weight fragments were found inthe RCC patients [47] Here we have shown an increase ofcfDNA concentration in RCC patients using genomic cfDNAfragment of 120573-actin gene of 99 bp Recent experiments fromother laboratories also demonstrated increased cfDNA levels
Disease Markers 7
in the blood of patientswith renal cancer compared to healthyindividuals [19 48 49]
Absolute values of cfDNAconcentrations obtained by twodistinct methods are different (11ndash2249 ngmL of plasma inthe fluorescence test compared to 23ndash1177 ngmL in qPCR)but in both cases they were significantly higher than inhealthy individuals (4ndash426 ngmL in the fluorescence testand 3ndash146 ngmL in qPCR) The obtained results agree withdata from other studies in which the determination of thecfDNA concentration using fluorescent dyes gave highervalues than qPCR [24 41] The high AUC values obtainedfor both methods of cfDNA concentration measurement inRCC patients (07679 for fluorescent test and 080494 forqPCR) demonstrate that these methods can be used forclinical investigations In addition to quantitative changescfDNA also possesses qualitative changes that occur in DNAof tumor cells such as mutations microsatellite instabilityand methylation [11 41 50] Methylation of gene promotersis a well-known mechanism of regulation of gene expression[51] Most frequently aberrantmethylation of genes occurs incancer cells Aberrant methylation of the promoter detectedin cfDNA can be used for noninvasive detection of cancerdifferential diagnosis prognosis of survival and response tocancer therapy [52ndash57] Currently several diagnostic systemsbased on the detection of DNA methylation exist that areaimed not only at detecting malignancy (MethylMeter fromRiboMed USA) but also at detecting specific types of cancer(Epi proColon and Epi proLung from Epigenomics AG Ger-many Confirm MDx for Prostate Cancer from MDxHealthUSA Product series DecisionDx G-CIMP Melanom ECUMThymoma from Castle Biosciences) The search for newtools is being pursued because only two of these systems(MethylMeter and Epi proColon) can detect cancer at earlystages of development and can monitor treatment since theyare based on the detection of cfDNA methylation
In this study we started investigating methylation ofpreviously identified tumor suppressor genes in cfDNAData from many studies show that the RASSF1 gene playsan important role in cancerogenesis Hypermethylation ofRASSF1 CpG islands is associated with different types ofcancer and with the risk of progression of tumorigenesis [58ndash64] It was also shown that rat RASSF1 is involved in earlytumorigenesis of RCC [16 17] Studies on methylation of thisgene in blood serum led to controversial results Hauser et al[18] showed that RASSF1A is methylated in 229 of patientsthe study of De Martino et al [19] demonstrated methylationof RASSF1A in 459 of patients Hoque et al [20] observedmethylation of this gene in 11 of serum samples of patientswith RCC In our study methylation of RASSF1 was detectedin 629 of patients The differences in methylation levels ofthe RASSF1 gene can be explained by the use of different CpGislands for analyses We studied methylation of CpG regionlocated within the first exon of RASSF1C while Hauser et al[18] and De Martino et al [19] analysed the region locatedupstream of the initiation codon Previously it was reportedthat these two CpG islands were differentially methylatedin melanoma cell lines and melanoma tumors [65] Ellingeret al [66] demonstrated a 100 correlation between DNAhypermethylation of the RASSF1A promoter and papillary
RCC However De Martino et al [19] analysed 31 samplesof papillary RCCs and found no association of RASSF1Amethylation with the histological subtypes of RCC In ourstudy RASSF1 was also methylated in papillary RCC but itwas the only sample of this cancer subtype analysed
Previously we reported changes in the LRRC3B genepromoter during the search for genetic and epigenetic alter-ations in chromosome 3 in epithelial tumors using NotI-microarrays [14 67 68] LRRC3B was identified by Kim etal [69] as a putative gene suppressor of several tumors thatare silenced in gastric cancers by epigenetic mechanismsIncreased methylation of the LRRC3B gene promoter wasconfirmed in samples of clear cell RCC and colorectalhead and neck cancer [29 70 71] A high level of LRRC3Bhypermethylation was noted not only in RCC patients (74)but also in healthy donors (33) in our study questioning theuse of this gene for the diagnosis of renal cancer on cfDNA
The promoter of the APC gene was methylated in 519of patients which is in good agreement with the results ofHauser and colleagues [18] who detected methylation of theAPC gene in 543 of patients using cfDNA
Previously a significant correlation between FHITexpression in clear cell renal carcinomas and patientsurvival was demonstrated [21] Kvasha et al [22] showeda correlation between hypermethylation of the FHIT CpGisland and a significant decrease of FHIT expression inclear cell RCC The level of aberrant methylation of FHITobtained in our study on cfDNA (556) was close to theresults obtained in the study of Kvasha et al in samples ofRCC tumors (546)
Integrin 12057291205731 plays an important role in various signaltransduction pathways that control proliferation migrationand differentiation of both normal (reviewed in [72 73])and cancer cells (reviewed in [74 75]) Downregulation ofITGA9 expression was observed in several cancer types [76ndash78] that could be caused either by mutations in this gene [79]or by hypermethylation [31 68 80] However methylationof the ITGA9 gene was not detected in our experimentsWe also have not identified methylation of the VHL genealthough NotI-microarray hybridization revealed high levelsof changes in this gene (47) in renal cancer [14] It is possiblethat these changes are associated with deletions in the generather than with methylation At the same time in the studyof DeMartino et al where cfDNAwas analysed by restrictionanalysis methylated VHL was detected in 503 of patientswith RCC [19]
Methylation analysis of the RASSF1 FHIT and APCgenes demonstrated their high specificity (933 for RASSF1and APC 100 for FHIT) for renal tumors Neverthelesssensitivity in one gene analysis was just from 519 forAPC to 629 for RASSF1 (Table 4) At the same timethe use of a combination of three or two genes (withoutLRRC3B due to the low specificity of this gene) leads to asignificant increase in sensitivity (779ndash923) and specificity(867ndash933) All other combinations did not reveal anyadditional diagnosis information Simultaneous methylationof the RASSF1 APC and FHIT genes was identified only in3 patients with metastases However the small sample sizedoes not allow us to draw a conclusion on the correlation
8 Disease Markers
between methylation and disease progression At the sametime binary logistic regression analysis showed the consid-erable diagnostic potential of combining both approachesused in this study According to the ROC analysis the use ofonly cfDNA concentration has moderate diagnostic potential(AUC = 08) On the other hand by using the concentrationand methylation of two or three genes we achieved 100diagnostic accuracy in our samples These results of coursecannot be directly transferred to clinical practice and needverification on a larger number of samples However our datademonstrates the potential advantage there is in combiningevaluation of cfDNA concentration and gene methylation forRCC diagnostics and provides a basis for further research
Thus despite the small sampling our results confirm thepossibility of using the concentration of cfDNA in bloodplasma as an additional marker of renal cancer developmentand show that methylation of three genes FHIT APC andRASSF1 in cfDNA can be used to develop renal cancerdiagnostic tools
5 Conclusion
The results obtained indicate that the concentration of cell-free DNA in plasma and the methylation of specific genes(such as FHIT APC and RASSF1) can be a significantaddition to serological tumor markers in the identification ofpatients with renal cancer However further studies need tobe performed to evaluate their diagnostic value
Competing Interests
The authors declare that they have no competing interests
Acknowledgments
The authors thank Dr A-L Haenni for helpful discussionsand comments on the manuscript This work was supportedby Grants 0110U004534 and 115U002951 from NationalAcademy of Sciences of Ukraine
References
[1] G Banumathy and P Cairns ldquoSignaling pathways in renal cellcarcinomardquo Cancer Biology andTherapy vol 10 no 7 pp 658ndash664 2010
[2] K Gupta J D Miller J Z Li M W Russell and C Charbon-neau ldquoEpidemiologic and socioeconomic burden of metastaticrenal cell carcinoma (mRCC) a literature reviewrdquo CancerTreatment Reviews vol 34 no 3 pp 193ndash205 2008
[3] Ministry of Healthcare of Ukraine The Features Diagnosis andPrognosis Factors Renal Cell Carcinoma Ministry of Healthcareof Ukraine 2011 httpwwwsouuorgua
[4] World Cancer Research Fund InternationalAmerican Institutefor Cancer Research Continuous Update Project Report DietNutrition Physical Activity and Kidney Cancer 2015
[5] A Lopez-Beltran M Scarpelli R Montironi and Z Kirkalildquo2004 WHO classification of the renal tumors of the adultsrdquoEuropean Urology vol 49 no 5 pp 798ndash805 2006
[6] P Hadaczek Z Siprashvili M Markiewski et al ldquoAbsenceor reduction of FHIT expression in most clear cell renalcarcinomasrdquo Cancer Research vol 58 no 14 pp 2946ndash29511998
[7] E Arai and Y Kanai ldquoGenetic and epigenetic alterationsduring renal carcinogenesisrdquo International Journal of Clinicaland Experimental Pathology vol 4 no 1 pp 58ndash73 2011
[8] B N Lasseigne T C Burwell M A Patil D M Absher J DBrooks and R M Myers ldquoDNA methylation profiling revealsnovel diagnostic biomarkers in renal cell carcinomardquo BMCMedicine vol 12 no 1 article 235 2014
[9] N Shenoy N Vallumsetla Y Zou et al ldquoRole of DNAmethylation in renal cell carcinomardquo Journal of Hematology andOncology vol 8 no 1 article 88 2015
[10] G J Fournie J-P Courtin F Laval et al ldquoPlasma DNA asa marker of cancerous cell death Investigations in patientssuffering from lung cancer and in nude mice bearing humantumoursrdquo Cancer Letters vol 91 no 2 pp 221ndash227 1995
[11] H Schwarzenbach D S B Hoon and K Pantel ldquoCell-freenucleic acids as biomarkers in cancer patientsrdquo Nature ReviewsCancer vol 11 no 6 pp 426ndash437 2011
[12] B Taback and D S B Hoon ldquoCirculating nucleic acids andproteomics of plasmaserum clinical utilityrdquo Annals of the NewYork Academy of Sciences vol 1022 pp 1ndash8 2004
[13] R Wasserkort A Kalmar G Valcz et al ldquoAberrant septin 9DNA methylation in colorectal cancer is restricted to a singleCpG islandrdquo BMC Cancer vol 13 article 398 2013
[14] I Ya Skrypkina V I Kashuba V V Gordiyuk et al ldquoIdentifi-cation of changes in gene loci potentially associated with renalcancer by novel technique of NotI microarraysrdquo Reports of theNational Academy of Sciences of Ukraine vol 11 pp 188ndash1922006
[15] V I Kashuba I Y Skrypkina D V Saraev et al ldquoIdentificationof changes in gene loci potentially associated with cervical can-cer usingNotImicroarraysrdquoUkrainrsquoskyi Biokhimichnyi Zhurnalvol 78 no 2 pp 113ndash120 2006
[16] V I Loginov D S Khodyrev I V Pronina et al ldquoMethylationof the RASSF1A promoter region and the allelic imbalancefrequencies in chromosome 3 critical regions correlate withprogression of clear cell renal carcinomardquo Molecular Biologyvol 43 no 3 pp 394ndash402 2009
[17] I Peters K Rehmet N Wilke et al ldquoRASSF1A promotermethylation and expression analysis in normal and neoplastickidney indicates a role in early tumorigenesisrdquo MolecularCancer vol 6 article 49 2007
[18] S Hauser T Zahalka and G Fechner ldquoSerum DNA hyperme-thylation in patients with kidney cancer results of a prospectivestudyrdquo Anticancer Research vol 33 pp 4651ndash4656 2013
[19] M De Martino T Klatte A Haitel and M Marberger ldquoSerumcell-free DNA in renal cell carcinoma a diagnostic and prog-nostic markerrdquo Cancer vol 118 no 1 pp 82ndash90 2012
[20] M O Hoque S Begum O Topaloglu et al ldquoQuantitativedetection of promoter hypermethylation of multiple genes inthe tumor urine and serumDNAof patients with renal cancerrdquoCancer Research vol 64 no 15 pp 5511ndash5517 2004
[21] U Ramp E Caliskan T Ebert et al ldquoFHIT expression inclear cell renal carcinomas versatility of protein levels andcorrelation with survivalrdquoThe Journal of Pathology vol 196 no4 pp 430ndash436 2002
[22] S Kvasha V Gordiyuk A Kondratov et al ldquoHypermethylationof the 51015840CpG island of the FHIT gene in clear cell renalcarcinomasrdquo Cancer Letters vol 265 no 2 pp 250ndash257 2008
Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

Disease Markers 3
are listed RASSF1 methylated-specific forward 51015840-GTG-TTAACGCGTTGCGTATC-31015840 and reverse 51015840-AACCCC-GCGAACTAAAAACGA-31015840 (60∘C 93 bp) [26] FHIT 51015840-TTGGGGCGCGGGTTTGGGTTTTTACGC-31015840 and 51015840-CGTAAACGACGCCGACCCCACTA-31015840 (62∘C 74 bp) [27]APC 51015840-TATTGCGGAGTGCGGGTC-31015840 and 51015840-TCGACG-AACTCCCGACGA-31015840 (60∘C 98 bp) [28] LRRC3B 51015840-GGTGCGAGGAAGGTAGGC-31015840 and 51015840-ACCAATACC-TCGCCGACG-31015840 (64∘C 149 bp) [29] VHL 51015840-TGGAGG-ATTTTTTTGCGTACGC-31015840 and 51015840-GAACCGAACGCC-GCGAA-31015840 (60∘C 158 bp) [30] ITGA9 51015840-TGGAGTATT-TTTACGATAATACGC-31015840 and 51015840-AAAAACCGAAAA-AACGACGA-31015840 (64∘C 116 bp) [31] Two 120583L of bisulfite-modified DNA was subjected to PCR amplification in a finalreaction volume of 25 120583L 1x Maxima SYBR Green qPCRMaster Mix (Cat number K0251 Thermo Scientific USA)and 03 120583M of each primer PCR was performed with aninitial 10min incubation at 95∘C followed by 45 cycles ofdenaturation at 95∘C for 15 s annealing for 20 s extensionat 72∘C for 30 s and a final 7min hold at 72∘C Each samplewas assayed in triplicate and each run included water blanksand an external control (universal methylated DNA) Afully methylated positive control was created by treating theDNA of lymphocytes from healthy donors with SssI CpGMethyltransferase (Cat number EM0821 Thermo ScientificUSA) according to themanufacturerrsquos recommendationsThespecificity of the PCR products was confirmed by meltingcurve analysis To verify MS-PCR data the MSP sequencingassay was performed using Genetic Analyser 3130 (AppliedBiosystems USA) following manufacturerrsquos protocols
26 Statistical Analysis Samples sizes were calculated usingthe formula described in [32] assuming 120572 and 120573 values of 005and 02 respectively We used standard deviation obtained inour preliminary experiments and estimated 150 differencein means
To evaluate the statistical significance of differencesbetween groups we performed the nonparametric Mann-Whitney 119880 test using the OriginPro 91 software (OriginLabUSA) or the Chi-square test (1205942) using Microsoft Excel 2007in the case of categorical variables
Differences were considered statistically significant if 119901 lt005 To evaluate the discriminative power of the parametersstudied for kidney cancer diagnostics we built binary logis-tic regression models for the selected predicting variablesand all their possible combinations using SPSS version 22(IBM USA) From these models the probabilities of positiveoutcome (ie cancer occurrence) were calculated Theseprobabilities were used for Receiver-operating characteristics(ROC) analysis Building of ROC and evaluation of AUC(Area Under Curve) was performed using the GraphPadPrism 607 (GraphPad Software La Jolla CA USA) or theOriginPro 91 software (OriginLab USA)
3 Results
31 Concentration of cfDNA in Blood Plasma of Patients withRenal Cancer and of Healthy Donors In this study bloodsamples from 27 patients with renal cancer and from 15
Table 1 Patient and tumor characteristics
Number of patientsAge at diagnosisAge gt 55 20 (741)Age lt 55 7 (259)
GenderMale 13 (481)Female 14 (519)
HistologyClear cell 23 (852)Sarcoma-like 2 (74)Papillary (75)clear cell (25) 1 (37)Cancer of the renal pelvis 1 (37)
Fuhrman gradeG1 + G2 19 (704)G3 + G4 8 (296)
Clinical stageStage 2 4 (148)Stage 3 23 (852)
Tumor-Node-Metastasis (TNM)T1a+b N0 M0-X 15 (556)T2 N0 M0-X 6 (222)T3 N0-1 M1-X 4 (148)TNM NA 2 (74)
healthy donors were used The blood samples were collectedbefore surgery in the Institute of Urology NAMS of UkraineThe results of the histological examination of tumors showedthat 23 patients had clear cell carcinoma 2 patients hadsarcoma-like tumors 1 patient had mixed type RCC (papil-laryclear cell) and 1 patient had cancer of the renal pelvis(Table 1)
The concentrations of cfDNA in blood plasma weredetermined by two methods by measuring the fluorescencelevel of intercalated SYBR Green I dye and by quantitativereal-time PCR (qPCR) of the 120573-actin gene
The results of the SYBR Green I fluorescence measure-ments showed that the concentrations of cfDNA in patientswith renal cancer range from 113 to 224912 ngmL (median23555 ngmL) The range of cfDNA concentration in healthydonors was much lower from 329 to 42675 ngmL (median5366 ngmL) (Figure 1(a))
qPCR revealed a statistically significant increaseof cfDNA concentration in cancer patients (median8097 ngmL range 233ndash11766 ngmL of plasma) As canbe seen from the box plot (Figure 1(c)) these values aresignificantly higher in RCC patients compared to healthydonors (median 351 ngmL 30ndash14678 ngmL of plasma)(Figure 1(c))
Receiver-operating characteristics (ROC) analysisshowed that the concentration of cfDNA can be used asdiagnostic feature for the detection of renal tumors (Figures1(b)ndash1(d)) AUC obtained for qRCR analysis was slightlyhigher (08049 119901 = 00012) than for the SYBR Green Ifluorescence measurements (07679 119901 = 00044) (Table 2)
4 Disease Markers
Table 2 Comparative analysis of different methods used to measure cfDNA in blood plasma
MethodqPCR analysis SYBR Green I fluorescence measurements
AUC 08049 (95 Cl 06602ndash09497) 07679 (95 Cl 06242ndash09116)Median (renal cancer) 8096 2355Median (control) 351 537119901 value (by Mann-Whitney 119880 test) 119901 lt 00008 119901 lt 00037
Renal cancer Control
0
1000
2000
Plas
ma D
NA
conc
entr
atio
n (n
gm
L)
p = 00037
(a)
0
50
100
Sens
itivi
ty (
)
50 1000100minus specificity ()
(b)
Renal cancer Control
p = 00008
0
500
1000
1500
Plas
ma D
NA
conc
entr
atio
n (n
gm
L)
(c)
50 10000
50
100
100minus specificity ()
Sens
itivi
ty (
)
(d)
Figure 1 Analysis of cfDNA concentration in plasma of patients with renal carcinoma and controls cfDNA concentrations were determinedby measuring the fluorescence level of intercalated SYBR Green I dye (a) and by qPCR (c) ROC curve analysis of cfDNA concentration incancer patients compared with the control group ((b) fluorescence test (d) qPCR)
32 Analysis of Methylation of Tumor Suppressor Genes incfDNA Since the cfDNA level alone cannot be a specificmarker of renal cancer [11] we also analysed the methylationstatus of CpG islands of 6 tumor suppressor genes in thecfDNA Using bisulfite treatment followed by MS-PCR wedetectedmethylation of theLRRC3BAPC FHIT andRASSF1genes in the cfDNA of cancer patients Promoter methylationof the LRRC3B gene was detected in 20 out of 27 samples(74) methylation of the RASSF1 APC and FHIT geneswas found in 17 (63) 14 (52) and 15 (556) patients
respectively (see Table 3 for detailed methylation frequen-cies)Methylation was not detected in the VHL and Integrin12057291205731 (ITGA9) genes in plasma cfDNA
Analysis of simultaneous methylation of CpG islands ofthe LRRC3B FHIT 0Pb and RASSF1 genes showed thatall the samples from cancer patients contained at least onemethylated promoter two promoters were methylated in333 three promoters were methylated in 27 and fourmethylated promoters were detected in 111 of the samples(Tables 3 and 4)
Disease Markers 5
Table 3 Summary of clinicopathological characteristics of patients with RCC and methylation status of LRRC3B RASSF1 FHIT and APCCpG islands in cfDNAlowast
Number Pathology Age (y) Sex pTNM Clinical grade Fuhrman nuclear grade MethylationLRRC3B RASSF1 APC FHIT
1 ccRCC 54 M af2N0M0 II 3 + + minus +2 ccRCC 61 M f1N0M0 II 2 + + minus +3 Sarcoma-like 66 F af2N0MX II 3 + minus + +4 PapillaryccRCC 63 M af1kN0MX III 1 minus + minus +5 ccRCC 47 F af31N0M1 III 3 + + + +6 ccRCC 64 M af31N0M1 III 3 + + + +7 ccRCC 58 M af2N0MX III 3 + minus + minus
8 ccRCC 61 M af1kN0M0 III 2 + + minus minus
9 ccRCC 75 M af1kN0MX III 2 minus + minus +10 ccRbb 65 M T2N0MX III 3 minus + + +11 ccRCC 61 F pT1N0M0 III 2 + minus + minus
12 ccRCC 63 F af31N1MX III 2-3 + minus + minus
13 ccRCC 68 F pT1 N0 MX III 1-2 + minus + +14 ccRCC 34 M af11N0 MX III 1 + minus + minus
15 Cancer of the renal pelvis 76 M pT3N0M1 III 4 + + + +16 ccRCC 56 F af11N0MX III 1 minus minus + +17 ccRCC 62 F af11N0MX III 1 + minus + +18 ccRCC 46 F af1kN0MX III 1 + + minus minus
19 ccRCC 55 F af2N0MX III 2 + + + minus
20 ccRCC 45 F T2N0M0 II 2 minus + minus minus
21 ccRCC 61 F af11N0MX III 2 + + minus minus
22 ccRCC 60 M NA III 2 + + minus +23 ccRCC 63 F af1kN0MX III 2 + minus minus minus
24 Sarcoma-like 60 F NA III 4 + minus minus minus
25 ccRCC 45 M pT1kN0MX III 2 minus + + +26 ccRCC 63 M af11N0M0 III 1 + + minus +27 ccRCC 73 F af11N0M0 III 1 minus + minus minus
lowastThe results in the Table are presented only for the genes with detected aberrant methylation in cfDNA
Table 4 Diagnostic data analysis for the discrimination of renal cancer patients and healthy subjects using cfDNA methylation of variousgenes alone and in combination
Markers Renal cell carcinoma (119899 = 27) Healthy controls (119899 = 15) 1205942 119901 value Sensitivitylowast Specificitylowastlowast LRRC3B 20 (741) 5 (333) 001 741 667RASSF1 17 (630 ) 1 (67) 00058 629 933FHIT 15 (556) 0 (0) 00003 556 100APC 14 (519) 1 (67) 00034 519 933VHL 0 (0) 0 (0) 0 100ITGA9 0 (0) 0 (0) 0 100RASSF1 or FHIT or APC 25 (923) 2 (133) lt00001 923 867RASSF1 or FHIT 21 (778) 1 (67) lt00001 778 933RASSF1 or APC 21 (778) 1 (67) lt00001 778 933lowastSensitivity was calculated as a percentage of positive results from a number of tested RCC patients lowastlowastspecificity was calculated as a percentage of negativetests from a given number of healthy donors
However LRRC3B showed a low specificity as a markerof cancer since it was methylated in 5 out of 15 (333)healthy donors Methylation of FHIT was not detected in thecfDNA of the control group while methylation of the APCand RASSF1 genes was found in 1 out of 15 (67) healthy
donors Methylation of APC and RASSF1 was detected indifferent healthy individuals (Table 4)
The sensitivity of each of thesemarkers exceeded 50 andwas 519 for APC 63 for RASSF1 and 556 for FHITwhich exhibited the best specificity in our test (100)
6 Disease Markers
Table 5 Receiver-operating characteristic (ROC) curve analyses of cfDNA marker-based models to discriminate between healthy persons(119899 = 15) and renal cancer patients (119899 = 27)lowast
AUC Std error 119901 value 95 LCL 95 UCLConc qPCR 080494 006771 000119 067223 093765Conc qPCR+APC 091852 004205 861E minus 06 08361 100094Conc qPCR+FHIT 091358 004898 110E minus 05 081759 100957Conc qPCR+RASSF1 088148 005986 500E minus 05 076416 099881Conc qPCR+ APC+RASSF1 1 0 106E minus 07 1 1Conc qPCR+APC+FHIT 095802 003018 112E minus 06 089888 101717Conc qPCR+RASSF1A+FHIT 094568 004521 216E minus 06 085708 103428Conc qPCR+APC+FHIT+RASSF1 1 0 106E minus 07 1 1lowastCalculated by binary logistic regression using combination of differentmarkers concentration of cfDNA determined by qPCR (Conc qPCR) andmethylationmarker genes (APC FHIT and RASSF1)
The use of the combined analysis of methylation statusof three genes (RASSF1 FHIT and APC) increased thesensitivity (778ndash923) while the specificity remained high(867ndash933) (Table 4) We did not find any correlationbetween hypermethylation cfDNA concentration and clin-icopathological parameters (grade lymph node metastasisage and sex) in patients with renal cancer
To explore the potential of combined cfDNA concen-tration and gene methylation for RCC diagnostics we per-formed binary logistic regression modelling As a predictorof variables we used cfDNA concentrations measured byquantitative PCR andmethylation ofAPC FHIT andRASSF1genes We built separate models for cfDNA concentrationalone and for all possible combinations of cfDNA concentra-tion and methylation of one two or three genes Predictiveproperties of the models were compared by ROC analysis Asreported above the AUC value for the cfDNA concentrationalone was 08 Addition of one of the genes slightly increasedthe AUC to values 088ndash0918 although these differenceswere not statistically significant as can be seen from 95confidence intervals (Table 5) Addition of two genes ledto further increase of the AUC value up to 1 when usingAPC and RASSF1 Finally the AUC value was 1 when weused the cfDNA concentration and methylation of all threegenes studied The results of ROC analysis are summarisedin Table 5 some of representative ROC curves are shown inFigure 2
4 Discussion
The level of cfDNA in blood plasma could be a universalmarker of malignancy [33] Many studies have shown thatchanges in cfDNA concentration can be correlated withdevelopment prognosis and survival of cancer patients Anincrease of cfDNA concentration was observed in patientswith breast gastric ovary lung colon and prostate cancer[11 34ndash39] It was suggested that an increase of cfDNAconcentration in cancer patients is associated with apoptosisand necrosis of cancer cells in the tumor microenvironment[40] This suggestion was supported by numerous cancer-specific alterations (such as allelic imbalances methylationand mutations) that were found in blood cfDNA (for reviews
00
02
04
06
08
10
Sens
itivi
ty
minus02 1000 02 06 0804
Conc_qPCR
Reference lineConc_qPCR APC+Conc_qPCR APC+ + FHIT
Conc_qPCR APC+ +FHIT + RASSF1A
1minus specificity
Figure 2 Receiver-operating characteristics (ROC) curves obtainedby using different models for the discrimination between healthycontrols (119899 = 15) and renal carcinoma patients (119899 = 27)Conc qPCR concentration of cfDNA determined by qPCR
see [11 41]) It was also demonstrated that monitoring of thecfDNA level in peripheral blood can be used as biomarker ofresponse to therapy in different cancer types [38 42 43]
Previous studies demonstrated that the evaluation ofconcentration of low molecular weight cfDNA (up to 100 bp)is the most representative for detection of malignancies anddisease prognosis since the level of fragments of this sizeincreases with disease progression [44ndash47] Recently Lu etal [47] showed that cfDNA fragments of 67 bp and 180 bpdid not differ between the controls and nonmetastatic RCCpatients while the cfDNA integrity index decreased fromcontrol to the metastatic group Significantly higher concen-trations of low molecular weight fragments were found inthe RCC patients [47] Here we have shown an increase ofcfDNA concentration in RCC patients using genomic cfDNAfragment of 120573-actin gene of 99 bp Recent experiments fromother laboratories also demonstrated increased cfDNA levels
Disease Markers 7
in the blood of patientswith renal cancer compared to healthyindividuals [19 48 49]
Absolute values of cfDNAconcentrations obtained by twodistinct methods are different (11ndash2249 ngmL of plasma inthe fluorescence test compared to 23ndash1177 ngmL in qPCR)but in both cases they were significantly higher than inhealthy individuals (4ndash426 ngmL in the fluorescence testand 3ndash146 ngmL in qPCR) The obtained results agree withdata from other studies in which the determination of thecfDNA concentration using fluorescent dyes gave highervalues than qPCR [24 41] The high AUC values obtainedfor both methods of cfDNA concentration measurement inRCC patients (07679 for fluorescent test and 080494 forqPCR) demonstrate that these methods can be used forclinical investigations In addition to quantitative changescfDNA also possesses qualitative changes that occur in DNAof tumor cells such as mutations microsatellite instabilityand methylation [11 41 50] Methylation of gene promotersis a well-known mechanism of regulation of gene expression[51] Most frequently aberrantmethylation of genes occurs incancer cells Aberrant methylation of the promoter detectedin cfDNA can be used for noninvasive detection of cancerdifferential diagnosis prognosis of survival and response tocancer therapy [52ndash57] Currently several diagnostic systemsbased on the detection of DNA methylation exist that areaimed not only at detecting malignancy (MethylMeter fromRiboMed USA) but also at detecting specific types of cancer(Epi proColon and Epi proLung from Epigenomics AG Ger-many Confirm MDx for Prostate Cancer from MDxHealthUSA Product series DecisionDx G-CIMP Melanom ECUMThymoma from Castle Biosciences) The search for newtools is being pursued because only two of these systems(MethylMeter and Epi proColon) can detect cancer at earlystages of development and can monitor treatment since theyare based on the detection of cfDNA methylation
In this study we started investigating methylation ofpreviously identified tumor suppressor genes in cfDNAData from many studies show that the RASSF1 gene playsan important role in cancerogenesis Hypermethylation ofRASSF1 CpG islands is associated with different types ofcancer and with the risk of progression of tumorigenesis [58ndash64] It was also shown that rat RASSF1 is involved in earlytumorigenesis of RCC [16 17] Studies on methylation of thisgene in blood serum led to controversial results Hauser et al[18] showed that RASSF1A is methylated in 229 of patientsthe study of De Martino et al [19] demonstrated methylationof RASSF1A in 459 of patients Hoque et al [20] observedmethylation of this gene in 11 of serum samples of patientswith RCC In our study methylation of RASSF1 was detectedin 629 of patients The differences in methylation levels ofthe RASSF1 gene can be explained by the use of different CpGislands for analyses We studied methylation of CpG regionlocated within the first exon of RASSF1C while Hauser et al[18] and De Martino et al [19] analysed the region locatedupstream of the initiation codon Previously it was reportedthat these two CpG islands were differentially methylatedin melanoma cell lines and melanoma tumors [65] Ellingeret al [66] demonstrated a 100 correlation between DNAhypermethylation of the RASSF1A promoter and papillary
RCC However De Martino et al [19] analysed 31 samplesof papillary RCCs and found no association of RASSF1Amethylation with the histological subtypes of RCC In ourstudy RASSF1 was also methylated in papillary RCC but itwas the only sample of this cancer subtype analysed
Previously we reported changes in the LRRC3B genepromoter during the search for genetic and epigenetic alter-ations in chromosome 3 in epithelial tumors using NotI-microarrays [14 67 68] LRRC3B was identified by Kim etal [69] as a putative gene suppressor of several tumors thatare silenced in gastric cancers by epigenetic mechanismsIncreased methylation of the LRRC3B gene promoter wasconfirmed in samples of clear cell RCC and colorectalhead and neck cancer [29 70 71] A high level of LRRC3Bhypermethylation was noted not only in RCC patients (74)but also in healthy donors (33) in our study questioning theuse of this gene for the diagnosis of renal cancer on cfDNA
The promoter of the APC gene was methylated in 519of patients which is in good agreement with the results ofHauser and colleagues [18] who detected methylation of theAPC gene in 543 of patients using cfDNA
Previously a significant correlation between FHITexpression in clear cell renal carcinomas and patientsurvival was demonstrated [21] Kvasha et al [22] showeda correlation between hypermethylation of the FHIT CpGisland and a significant decrease of FHIT expression inclear cell RCC The level of aberrant methylation of FHITobtained in our study on cfDNA (556) was close to theresults obtained in the study of Kvasha et al in samples ofRCC tumors (546)
Integrin 12057291205731 plays an important role in various signaltransduction pathways that control proliferation migrationand differentiation of both normal (reviewed in [72 73])and cancer cells (reviewed in [74 75]) Downregulation ofITGA9 expression was observed in several cancer types [76ndash78] that could be caused either by mutations in this gene [79]or by hypermethylation [31 68 80] However methylationof the ITGA9 gene was not detected in our experimentsWe also have not identified methylation of the VHL genealthough NotI-microarray hybridization revealed high levelsof changes in this gene (47) in renal cancer [14] It is possiblethat these changes are associated with deletions in the generather than with methylation At the same time in the studyof DeMartino et al where cfDNAwas analysed by restrictionanalysis methylated VHL was detected in 503 of patientswith RCC [19]
Methylation analysis of the RASSF1 FHIT and APCgenes demonstrated their high specificity (933 for RASSF1and APC 100 for FHIT) for renal tumors Neverthelesssensitivity in one gene analysis was just from 519 forAPC to 629 for RASSF1 (Table 4) At the same timethe use of a combination of three or two genes (withoutLRRC3B due to the low specificity of this gene) leads to asignificant increase in sensitivity (779ndash923) and specificity(867ndash933) All other combinations did not reveal anyadditional diagnosis information Simultaneous methylationof the RASSF1 APC and FHIT genes was identified only in3 patients with metastases However the small sample sizedoes not allow us to draw a conclusion on the correlation
8 Disease Markers
between methylation and disease progression At the sametime binary logistic regression analysis showed the consid-erable diagnostic potential of combining both approachesused in this study According to the ROC analysis the use ofonly cfDNA concentration has moderate diagnostic potential(AUC = 08) On the other hand by using the concentrationand methylation of two or three genes we achieved 100diagnostic accuracy in our samples These results of coursecannot be directly transferred to clinical practice and needverification on a larger number of samples However our datademonstrates the potential advantage there is in combiningevaluation of cfDNA concentration and gene methylation forRCC diagnostics and provides a basis for further research
Thus despite the small sampling our results confirm thepossibility of using the concentration of cfDNA in bloodplasma as an additional marker of renal cancer developmentand show that methylation of three genes FHIT APC andRASSF1 in cfDNA can be used to develop renal cancerdiagnostic tools
5 Conclusion
The results obtained indicate that the concentration of cell-free DNA in plasma and the methylation of specific genes(such as FHIT APC and RASSF1) can be a significantaddition to serological tumor markers in the identification ofpatients with renal cancer However further studies need tobe performed to evaluate their diagnostic value
Competing Interests
The authors declare that they have no competing interests
Acknowledgments
The authors thank Dr A-L Haenni for helpful discussionsand comments on the manuscript This work was supportedby Grants 0110U004534 and 115U002951 from NationalAcademy of Sciences of Ukraine
References
[1] G Banumathy and P Cairns ldquoSignaling pathways in renal cellcarcinomardquo Cancer Biology andTherapy vol 10 no 7 pp 658ndash664 2010
[2] K Gupta J D Miller J Z Li M W Russell and C Charbon-neau ldquoEpidemiologic and socioeconomic burden of metastaticrenal cell carcinoma (mRCC) a literature reviewrdquo CancerTreatment Reviews vol 34 no 3 pp 193ndash205 2008
[3] Ministry of Healthcare of Ukraine The Features Diagnosis andPrognosis Factors Renal Cell Carcinoma Ministry of Healthcareof Ukraine 2011 httpwwwsouuorgua
[4] World Cancer Research Fund InternationalAmerican Institutefor Cancer Research Continuous Update Project Report DietNutrition Physical Activity and Kidney Cancer 2015
[5] A Lopez-Beltran M Scarpelli R Montironi and Z Kirkalildquo2004 WHO classification of the renal tumors of the adultsrdquoEuropean Urology vol 49 no 5 pp 798ndash805 2006
[6] P Hadaczek Z Siprashvili M Markiewski et al ldquoAbsenceor reduction of FHIT expression in most clear cell renalcarcinomasrdquo Cancer Research vol 58 no 14 pp 2946ndash29511998
[7] E Arai and Y Kanai ldquoGenetic and epigenetic alterationsduring renal carcinogenesisrdquo International Journal of Clinicaland Experimental Pathology vol 4 no 1 pp 58ndash73 2011
[8] B N Lasseigne T C Burwell M A Patil D M Absher J DBrooks and R M Myers ldquoDNA methylation profiling revealsnovel diagnostic biomarkers in renal cell carcinomardquo BMCMedicine vol 12 no 1 article 235 2014
[9] N Shenoy N Vallumsetla Y Zou et al ldquoRole of DNAmethylation in renal cell carcinomardquo Journal of Hematology andOncology vol 8 no 1 article 88 2015
[10] G J Fournie J-P Courtin F Laval et al ldquoPlasma DNA asa marker of cancerous cell death Investigations in patientssuffering from lung cancer and in nude mice bearing humantumoursrdquo Cancer Letters vol 91 no 2 pp 221ndash227 1995
[11] H Schwarzenbach D S B Hoon and K Pantel ldquoCell-freenucleic acids as biomarkers in cancer patientsrdquo Nature ReviewsCancer vol 11 no 6 pp 426ndash437 2011
[12] B Taback and D S B Hoon ldquoCirculating nucleic acids andproteomics of plasmaserum clinical utilityrdquo Annals of the NewYork Academy of Sciences vol 1022 pp 1ndash8 2004
[13] R Wasserkort A Kalmar G Valcz et al ldquoAberrant septin 9DNA methylation in colorectal cancer is restricted to a singleCpG islandrdquo BMC Cancer vol 13 article 398 2013
[14] I Ya Skrypkina V I Kashuba V V Gordiyuk et al ldquoIdentifi-cation of changes in gene loci potentially associated with renalcancer by novel technique of NotI microarraysrdquo Reports of theNational Academy of Sciences of Ukraine vol 11 pp 188ndash1922006
[15] V I Kashuba I Y Skrypkina D V Saraev et al ldquoIdentificationof changes in gene loci potentially associated with cervical can-cer usingNotImicroarraysrdquoUkrainrsquoskyi Biokhimichnyi Zhurnalvol 78 no 2 pp 113ndash120 2006
[16] V I Loginov D S Khodyrev I V Pronina et al ldquoMethylationof the RASSF1A promoter region and the allelic imbalancefrequencies in chromosome 3 critical regions correlate withprogression of clear cell renal carcinomardquo Molecular Biologyvol 43 no 3 pp 394ndash402 2009
[17] I Peters K Rehmet N Wilke et al ldquoRASSF1A promotermethylation and expression analysis in normal and neoplastickidney indicates a role in early tumorigenesisrdquo MolecularCancer vol 6 article 49 2007
[18] S Hauser T Zahalka and G Fechner ldquoSerum DNA hyperme-thylation in patients with kidney cancer results of a prospectivestudyrdquo Anticancer Research vol 33 pp 4651ndash4656 2013
[19] M De Martino T Klatte A Haitel and M Marberger ldquoSerumcell-free DNA in renal cell carcinoma a diagnostic and prog-nostic markerrdquo Cancer vol 118 no 1 pp 82ndash90 2012
[20] M O Hoque S Begum O Topaloglu et al ldquoQuantitativedetection of promoter hypermethylation of multiple genes inthe tumor urine and serumDNAof patients with renal cancerrdquoCancer Research vol 64 no 15 pp 5511ndash5517 2004
[21] U Ramp E Caliskan T Ebert et al ldquoFHIT expression inclear cell renal carcinomas versatility of protein levels andcorrelation with survivalrdquoThe Journal of Pathology vol 196 no4 pp 430ndash436 2002
[22] S Kvasha V Gordiyuk A Kondratov et al ldquoHypermethylationof the 51015840CpG island of the FHIT gene in clear cell renalcarcinomasrdquo Cancer Letters vol 265 no 2 pp 250ndash257 2008
Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

4 Disease Markers
Table 2 Comparative analysis of different methods used to measure cfDNA in blood plasma
MethodqPCR analysis SYBR Green I fluorescence measurements
AUC 08049 (95 Cl 06602ndash09497) 07679 (95 Cl 06242ndash09116)Median (renal cancer) 8096 2355Median (control) 351 537119901 value (by Mann-Whitney 119880 test) 119901 lt 00008 119901 lt 00037
Renal cancer Control
0
1000
2000
Plas
ma D
NA
conc
entr
atio
n (n
gm
L)
p = 00037
(a)
0
50
100
Sens
itivi
ty (
)
50 1000100minus specificity ()
(b)
Renal cancer Control
p = 00008
0
500
1000
1500
Plas
ma D
NA
conc
entr
atio
n (n
gm
L)
(c)
50 10000
50
100
100minus specificity ()
Sens
itivi
ty (
)
(d)
Figure 1 Analysis of cfDNA concentration in plasma of patients with renal carcinoma and controls cfDNA concentrations were determinedby measuring the fluorescence level of intercalated SYBR Green I dye (a) and by qPCR (c) ROC curve analysis of cfDNA concentration incancer patients compared with the control group ((b) fluorescence test (d) qPCR)
32 Analysis of Methylation of Tumor Suppressor Genes incfDNA Since the cfDNA level alone cannot be a specificmarker of renal cancer [11] we also analysed the methylationstatus of CpG islands of 6 tumor suppressor genes in thecfDNA Using bisulfite treatment followed by MS-PCR wedetectedmethylation of theLRRC3BAPC FHIT andRASSF1genes in the cfDNA of cancer patients Promoter methylationof the LRRC3B gene was detected in 20 out of 27 samples(74) methylation of the RASSF1 APC and FHIT geneswas found in 17 (63) 14 (52) and 15 (556) patients
respectively (see Table 3 for detailed methylation frequen-cies)Methylation was not detected in the VHL and Integrin12057291205731 (ITGA9) genes in plasma cfDNA
Analysis of simultaneous methylation of CpG islands ofthe LRRC3B FHIT 0Pb and RASSF1 genes showed thatall the samples from cancer patients contained at least onemethylated promoter two promoters were methylated in333 three promoters were methylated in 27 and fourmethylated promoters were detected in 111 of the samples(Tables 3 and 4)
Disease Markers 5
Table 3 Summary of clinicopathological characteristics of patients with RCC and methylation status of LRRC3B RASSF1 FHIT and APCCpG islands in cfDNAlowast
Number Pathology Age (y) Sex pTNM Clinical grade Fuhrman nuclear grade MethylationLRRC3B RASSF1 APC FHIT
1 ccRCC 54 M af2N0M0 II 3 + + minus +2 ccRCC 61 M f1N0M0 II 2 + + minus +3 Sarcoma-like 66 F af2N0MX II 3 + minus + +4 PapillaryccRCC 63 M af1kN0MX III 1 minus + minus +5 ccRCC 47 F af31N0M1 III 3 + + + +6 ccRCC 64 M af31N0M1 III 3 + + + +7 ccRCC 58 M af2N0MX III 3 + minus + minus
8 ccRCC 61 M af1kN0M0 III 2 + + minus minus
9 ccRCC 75 M af1kN0MX III 2 minus + minus +10 ccRbb 65 M T2N0MX III 3 minus + + +11 ccRCC 61 F pT1N0M0 III 2 + minus + minus
12 ccRCC 63 F af31N1MX III 2-3 + minus + minus
13 ccRCC 68 F pT1 N0 MX III 1-2 + minus + +14 ccRCC 34 M af11N0 MX III 1 + minus + minus
15 Cancer of the renal pelvis 76 M pT3N0M1 III 4 + + + +16 ccRCC 56 F af11N0MX III 1 minus minus + +17 ccRCC 62 F af11N0MX III 1 + minus + +18 ccRCC 46 F af1kN0MX III 1 + + minus minus
19 ccRCC 55 F af2N0MX III 2 + + + minus
20 ccRCC 45 F T2N0M0 II 2 minus + minus minus
21 ccRCC 61 F af11N0MX III 2 + + minus minus
22 ccRCC 60 M NA III 2 + + minus +23 ccRCC 63 F af1kN0MX III 2 + minus minus minus
24 Sarcoma-like 60 F NA III 4 + minus minus minus
25 ccRCC 45 M pT1kN0MX III 2 minus + + +26 ccRCC 63 M af11N0M0 III 1 + + minus +27 ccRCC 73 F af11N0M0 III 1 minus + minus minus
lowastThe results in the Table are presented only for the genes with detected aberrant methylation in cfDNA
Table 4 Diagnostic data analysis for the discrimination of renal cancer patients and healthy subjects using cfDNA methylation of variousgenes alone and in combination
Markers Renal cell carcinoma (119899 = 27) Healthy controls (119899 = 15) 1205942 119901 value Sensitivitylowast Specificitylowastlowast LRRC3B 20 (741) 5 (333) 001 741 667RASSF1 17 (630 ) 1 (67) 00058 629 933FHIT 15 (556) 0 (0) 00003 556 100APC 14 (519) 1 (67) 00034 519 933VHL 0 (0) 0 (0) 0 100ITGA9 0 (0) 0 (0) 0 100RASSF1 or FHIT or APC 25 (923) 2 (133) lt00001 923 867RASSF1 or FHIT 21 (778) 1 (67) lt00001 778 933RASSF1 or APC 21 (778) 1 (67) lt00001 778 933lowastSensitivity was calculated as a percentage of positive results from a number of tested RCC patients lowastlowastspecificity was calculated as a percentage of negativetests from a given number of healthy donors
However LRRC3B showed a low specificity as a markerof cancer since it was methylated in 5 out of 15 (333)healthy donors Methylation of FHIT was not detected in thecfDNA of the control group while methylation of the APCand RASSF1 genes was found in 1 out of 15 (67) healthy
donors Methylation of APC and RASSF1 was detected indifferent healthy individuals (Table 4)
The sensitivity of each of thesemarkers exceeded 50 andwas 519 for APC 63 for RASSF1 and 556 for FHITwhich exhibited the best specificity in our test (100)
6 Disease Markers
Table 5 Receiver-operating characteristic (ROC) curve analyses of cfDNA marker-based models to discriminate between healthy persons(119899 = 15) and renal cancer patients (119899 = 27)lowast
AUC Std error 119901 value 95 LCL 95 UCLConc qPCR 080494 006771 000119 067223 093765Conc qPCR+APC 091852 004205 861E minus 06 08361 100094Conc qPCR+FHIT 091358 004898 110E minus 05 081759 100957Conc qPCR+RASSF1 088148 005986 500E minus 05 076416 099881Conc qPCR+ APC+RASSF1 1 0 106E minus 07 1 1Conc qPCR+APC+FHIT 095802 003018 112E minus 06 089888 101717Conc qPCR+RASSF1A+FHIT 094568 004521 216E minus 06 085708 103428Conc qPCR+APC+FHIT+RASSF1 1 0 106E minus 07 1 1lowastCalculated by binary logistic regression using combination of differentmarkers concentration of cfDNA determined by qPCR (Conc qPCR) andmethylationmarker genes (APC FHIT and RASSF1)
The use of the combined analysis of methylation statusof three genes (RASSF1 FHIT and APC) increased thesensitivity (778ndash923) while the specificity remained high(867ndash933) (Table 4) We did not find any correlationbetween hypermethylation cfDNA concentration and clin-icopathological parameters (grade lymph node metastasisage and sex) in patients with renal cancer
To explore the potential of combined cfDNA concen-tration and gene methylation for RCC diagnostics we per-formed binary logistic regression modelling As a predictorof variables we used cfDNA concentrations measured byquantitative PCR andmethylation ofAPC FHIT andRASSF1genes We built separate models for cfDNA concentrationalone and for all possible combinations of cfDNA concentra-tion and methylation of one two or three genes Predictiveproperties of the models were compared by ROC analysis Asreported above the AUC value for the cfDNA concentrationalone was 08 Addition of one of the genes slightly increasedthe AUC to values 088ndash0918 although these differenceswere not statistically significant as can be seen from 95confidence intervals (Table 5) Addition of two genes ledto further increase of the AUC value up to 1 when usingAPC and RASSF1 Finally the AUC value was 1 when weused the cfDNA concentration and methylation of all threegenes studied The results of ROC analysis are summarisedin Table 5 some of representative ROC curves are shown inFigure 2
4 Discussion
The level of cfDNA in blood plasma could be a universalmarker of malignancy [33] Many studies have shown thatchanges in cfDNA concentration can be correlated withdevelopment prognosis and survival of cancer patients Anincrease of cfDNA concentration was observed in patientswith breast gastric ovary lung colon and prostate cancer[11 34ndash39] It was suggested that an increase of cfDNAconcentration in cancer patients is associated with apoptosisand necrosis of cancer cells in the tumor microenvironment[40] This suggestion was supported by numerous cancer-specific alterations (such as allelic imbalances methylationand mutations) that were found in blood cfDNA (for reviews
00
02
04
06
08
10
Sens
itivi
ty
minus02 1000 02 06 0804
Conc_qPCR
Reference lineConc_qPCR APC+Conc_qPCR APC+ + FHIT
Conc_qPCR APC+ +FHIT + RASSF1A
1minus specificity
Figure 2 Receiver-operating characteristics (ROC) curves obtainedby using different models for the discrimination between healthycontrols (119899 = 15) and renal carcinoma patients (119899 = 27)Conc qPCR concentration of cfDNA determined by qPCR
see [11 41]) It was also demonstrated that monitoring of thecfDNA level in peripheral blood can be used as biomarker ofresponse to therapy in different cancer types [38 42 43]
Previous studies demonstrated that the evaluation ofconcentration of low molecular weight cfDNA (up to 100 bp)is the most representative for detection of malignancies anddisease prognosis since the level of fragments of this sizeincreases with disease progression [44ndash47] Recently Lu etal [47] showed that cfDNA fragments of 67 bp and 180 bpdid not differ between the controls and nonmetastatic RCCpatients while the cfDNA integrity index decreased fromcontrol to the metastatic group Significantly higher concen-trations of low molecular weight fragments were found inthe RCC patients [47] Here we have shown an increase ofcfDNA concentration in RCC patients using genomic cfDNAfragment of 120573-actin gene of 99 bp Recent experiments fromother laboratories also demonstrated increased cfDNA levels
Disease Markers 7
in the blood of patientswith renal cancer compared to healthyindividuals [19 48 49]
Absolute values of cfDNAconcentrations obtained by twodistinct methods are different (11ndash2249 ngmL of plasma inthe fluorescence test compared to 23ndash1177 ngmL in qPCR)but in both cases they were significantly higher than inhealthy individuals (4ndash426 ngmL in the fluorescence testand 3ndash146 ngmL in qPCR) The obtained results agree withdata from other studies in which the determination of thecfDNA concentration using fluorescent dyes gave highervalues than qPCR [24 41] The high AUC values obtainedfor both methods of cfDNA concentration measurement inRCC patients (07679 for fluorescent test and 080494 forqPCR) demonstrate that these methods can be used forclinical investigations In addition to quantitative changescfDNA also possesses qualitative changes that occur in DNAof tumor cells such as mutations microsatellite instabilityand methylation [11 41 50] Methylation of gene promotersis a well-known mechanism of regulation of gene expression[51] Most frequently aberrantmethylation of genes occurs incancer cells Aberrant methylation of the promoter detectedin cfDNA can be used for noninvasive detection of cancerdifferential diagnosis prognosis of survival and response tocancer therapy [52ndash57] Currently several diagnostic systemsbased on the detection of DNA methylation exist that areaimed not only at detecting malignancy (MethylMeter fromRiboMed USA) but also at detecting specific types of cancer(Epi proColon and Epi proLung from Epigenomics AG Ger-many Confirm MDx for Prostate Cancer from MDxHealthUSA Product series DecisionDx G-CIMP Melanom ECUMThymoma from Castle Biosciences) The search for newtools is being pursued because only two of these systems(MethylMeter and Epi proColon) can detect cancer at earlystages of development and can monitor treatment since theyare based on the detection of cfDNA methylation
In this study we started investigating methylation ofpreviously identified tumor suppressor genes in cfDNAData from many studies show that the RASSF1 gene playsan important role in cancerogenesis Hypermethylation ofRASSF1 CpG islands is associated with different types ofcancer and with the risk of progression of tumorigenesis [58ndash64] It was also shown that rat RASSF1 is involved in earlytumorigenesis of RCC [16 17] Studies on methylation of thisgene in blood serum led to controversial results Hauser et al[18] showed that RASSF1A is methylated in 229 of patientsthe study of De Martino et al [19] demonstrated methylationof RASSF1A in 459 of patients Hoque et al [20] observedmethylation of this gene in 11 of serum samples of patientswith RCC In our study methylation of RASSF1 was detectedin 629 of patients The differences in methylation levels ofthe RASSF1 gene can be explained by the use of different CpGislands for analyses We studied methylation of CpG regionlocated within the first exon of RASSF1C while Hauser et al[18] and De Martino et al [19] analysed the region locatedupstream of the initiation codon Previously it was reportedthat these two CpG islands were differentially methylatedin melanoma cell lines and melanoma tumors [65] Ellingeret al [66] demonstrated a 100 correlation between DNAhypermethylation of the RASSF1A promoter and papillary
RCC However De Martino et al [19] analysed 31 samplesof papillary RCCs and found no association of RASSF1Amethylation with the histological subtypes of RCC In ourstudy RASSF1 was also methylated in papillary RCC but itwas the only sample of this cancer subtype analysed
Previously we reported changes in the LRRC3B genepromoter during the search for genetic and epigenetic alter-ations in chromosome 3 in epithelial tumors using NotI-microarrays [14 67 68] LRRC3B was identified by Kim etal [69] as a putative gene suppressor of several tumors thatare silenced in gastric cancers by epigenetic mechanismsIncreased methylation of the LRRC3B gene promoter wasconfirmed in samples of clear cell RCC and colorectalhead and neck cancer [29 70 71] A high level of LRRC3Bhypermethylation was noted not only in RCC patients (74)but also in healthy donors (33) in our study questioning theuse of this gene for the diagnosis of renal cancer on cfDNA
The promoter of the APC gene was methylated in 519of patients which is in good agreement with the results ofHauser and colleagues [18] who detected methylation of theAPC gene in 543 of patients using cfDNA
Previously a significant correlation between FHITexpression in clear cell renal carcinomas and patientsurvival was demonstrated [21] Kvasha et al [22] showeda correlation between hypermethylation of the FHIT CpGisland and a significant decrease of FHIT expression inclear cell RCC The level of aberrant methylation of FHITobtained in our study on cfDNA (556) was close to theresults obtained in the study of Kvasha et al in samples ofRCC tumors (546)
Integrin 12057291205731 plays an important role in various signaltransduction pathways that control proliferation migrationand differentiation of both normal (reviewed in [72 73])and cancer cells (reviewed in [74 75]) Downregulation ofITGA9 expression was observed in several cancer types [76ndash78] that could be caused either by mutations in this gene [79]or by hypermethylation [31 68 80] However methylationof the ITGA9 gene was not detected in our experimentsWe also have not identified methylation of the VHL genealthough NotI-microarray hybridization revealed high levelsof changes in this gene (47) in renal cancer [14] It is possiblethat these changes are associated with deletions in the generather than with methylation At the same time in the studyof DeMartino et al where cfDNAwas analysed by restrictionanalysis methylated VHL was detected in 503 of patientswith RCC [19]
Methylation analysis of the RASSF1 FHIT and APCgenes demonstrated their high specificity (933 for RASSF1and APC 100 for FHIT) for renal tumors Neverthelesssensitivity in one gene analysis was just from 519 forAPC to 629 for RASSF1 (Table 4) At the same timethe use of a combination of three or two genes (withoutLRRC3B due to the low specificity of this gene) leads to asignificant increase in sensitivity (779ndash923) and specificity(867ndash933) All other combinations did not reveal anyadditional diagnosis information Simultaneous methylationof the RASSF1 APC and FHIT genes was identified only in3 patients with metastases However the small sample sizedoes not allow us to draw a conclusion on the correlation
8 Disease Markers
between methylation and disease progression At the sametime binary logistic regression analysis showed the consid-erable diagnostic potential of combining both approachesused in this study According to the ROC analysis the use ofonly cfDNA concentration has moderate diagnostic potential(AUC = 08) On the other hand by using the concentrationand methylation of two or three genes we achieved 100diagnostic accuracy in our samples These results of coursecannot be directly transferred to clinical practice and needverification on a larger number of samples However our datademonstrates the potential advantage there is in combiningevaluation of cfDNA concentration and gene methylation forRCC diagnostics and provides a basis for further research
Thus despite the small sampling our results confirm thepossibility of using the concentration of cfDNA in bloodplasma as an additional marker of renal cancer developmentand show that methylation of three genes FHIT APC andRASSF1 in cfDNA can be used to develop renal cancerdiagnostic tools
5 Conclusion
The results obtained indicate that the concentration of cell-free DNA in plasma and the methylation of specific genes(such as FHIT APC and RASSF1) can be a significantaddition to serological tumor markers in the identification ofpatients with renal cancer However further studies need tobe performed to evaluate their diagnostic value
Competing Interests
The authors declare that they have no competing interests
Acknowledgments
The authors thank Dr A-L Haenni for helpful discussionsand comments on the manuscript This work was supportedby Grants 0110U004534 and 115U002951 from NationalAcademy of Sciences of Ukraine
References
[1] G Banumathy and P Cairns ldquoSignaling pathways in renal cellcarcinomardquo Cancer Biology andTherapy vol 10 no 7 pp 658ndash664 2010
[2] K Gupta J D Miller J Z Li M W Russell and C Charbon-neau ldquoEpidemiologic and socioeconomic burden of metastaticrenal cell carcinoma (mRCC) a literature reviewrdquo CancerTreatment Reviews vol 34 no 3 pp 193ndash205 2008
[3] Ministry of Healthcare of Ukraine The Features Diagnosis andPrognosis Factors Renal Cell Carcinoma Ministry of Healthcareof Ukraine 2011 httpwwwsouuorgua
[4] World Cancer Research Fund InternationalAmerican Institutefor Cancer Research Continuous Update Project Report DietNutrition Physical Activity and Kidney Cancer 2015
[5] A Lopez-Beltran M Scarpelli R Montironi and Z Kirkalildquo2004 WHO classification of the renal tumors of the adultsrdquoEuropean Urology vol 49 no 5 pp 798ndash805 2006
[6] P Hadaczek Z Siprashvili M Markiewski et al ldquoAbsenceor reduction of FHIT expression in most clear cell renalcarcinomasrdquo Cancer Research vol 58 no 14 pp 2946ndash29511998
[7] E Arai and Y Kanai ldquoGenetic and epigenetic alterationsduring renal carcinogenesisrdquo International Journal of Clinicaland Experimental Pathology vol 4 no 1 pp 58ndash73 2011
[8] B N Lasseigne T C Burwell M A Patil D M Absher J DBrooks and R M Myers ldquoDNA methylation profiling revealsnovel diagnostic biomarkers in renal cell carcinomardquo BMCMedicine vol 12 no 1 article 235 2014
[9] N Shenoy N Vallumsetla Y Zou et al ldquoRole of DNAmethylation in renal cell carcinomardquo Journal of Hematology andOncology vol 8 no 1 article 88 2015
[10] G J Fournie J-P Courtin F Laval et al ldquoPlasma DNA asa marker of cancerous cell death Investigations in patientssuffering from lung cancer and in nude mice bearing humantumoursrdquo Cancer Letters vol 91 no 2 pp 221ndash227 1995
[11] H Schwarzenbach D S B Hoon and K Pantel ldquoCell-freenucleic acids as biomarkers in cancer patientsrdquo Nature ReviewsCancer vol 11 no 6 pp 426ndash437 2011
[12] B Taback and D S B Hoon ldquoCirculating nucleic acids andproteomics of plasmaserum clinical utilityrdquo Annals of the NewYork Academy of Sciences vol 1022 pp 1ndash8 2004
[13] R Wasserkort A Kalmar G Valcz et al ldquoAberrant septin 9DNA methylation in colorectal cancer is restricted to a singleCpG islandrdquo BMC Cancer vol 13 article 398 2013
[14] I Ya Skrypkina V I Kashuba V V Gordiyuk et al ldquoIdentifi-cation of changes in gene loci potentially associated with renalcancer by novel technique of NotI microarraysrdquo Reports of theNational Academy of Sciences of Ukraine vol 11 pp 188ndash1922006
[15] V I Kashuba I Y Skrypkina D V Saraev et al ldquoIdentificationof changes in gene loci potentially associated with cervical can-cer usingNotImicroarraysrdquoUkrainrsquoskyi Biokhimichnyi Zhurnalvol 78 no 2 pp 113ndash120 2006
[16] V I Loginov D S Khodyrev I V Pronina et al ldquoMethylationof the RASSF1A promoter region and the allelic imbalancefrequencies in chromosome 3 critical regions correlate withprogression of clear cell renal carcinomardquo Molecular Biologyvol 43 no 3 pp 394ndash402 2009
[17] I Peters K Rehmet N Wilke et al ldquoRASSF1A promotermethylation and expression analysis in normal and neoplastickidney indicates a role in early tumorigenesisrdquo MolecularCancer vol 6 article 49 2007
[18] S Hauser T Zahalka and G Fechner ldquoSerum DNA hyperme-thylation in patients with kidney cancer results of a prospectivestudyrdquo Anticancer Research vol 33 pp 4651ndash4656 2013
[19] M De Martino T Klatte A Haitel and M Marberger ldquoSerumcell-free DNA in renal cell carcinoma a diagnostic and prog-nostic markerrdquo Cancer vol 118 no 1 pp 82ndash90 2012
[20] M O Hoque S Begum O Topaloglu et al ldquoQuantitativedetection of promoter hypermethylation of multiple genes inthe tumor urine and serumDNAof patients with renal cancerrdquoCancer Research vol 64 no 15 pp 5511ndash5517 2004
[21] U Ramp E Caliskan T Ebert et al ldquoFHIT expression inclear cell renal carcinomas versatility of protein levels andcorrelation with survivalrdquoThe Journal of Pathology vol 196 no4 pp 430ndash436 2002
[22] S Kvasha V Gordiyuk A Kondratov et al ldquoHypermethylationof the 51015840CpG island of the FHIT gene in clear cell renalcarcinomasrdquo Cancer Letters vol 265 no 2 pp 250ndash257 2008
Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

Disease Markers 5
Table 3 Summary of clinicopathological characteristics of patients with RCC and methylation status of LRRC3B RASSF1 FHIT and APCCpG islands in cfDNAlowast
Number Pathology Age (y) Sex pTNM Clinical grade Fuhrman nuclear grade MethylationLRRC3B RASSF1 APC FHIT
1 ccRCC 54 M af2N0M0 II 3 + + minus +2 ccRCC 61 M f1N0M0 II 2 + + minus +3 Sarcoma-like 66 F af2N0MX II 3 + minus + +4 PapillaryccRCC 63 M af1kN0MX III 1 minus + minus +5 ccRCC 47 F af31N0M1 III 3 + + + +6 ccRCC 64 M af31N0M1 III 3 + + + +7 ccRCC 58 M af2N0MX III 3 + minus + minus
8 ccRCC 61 M af1kN0M0 III 2 + + minus minus
9 ccRCC 75 M af1kN0MX III 2 minus + minus +10 ccRbb 65 M T2N0MX III 3 minus + + +11 ccRCC 61 F pT1N0M0 III 2 + minus + minus
12 ccRCC 63 F af31N1MX III 2-3 + minus + minus
13 ccRCC 68 F pT1 N0 MX III 1-2 + minus + +14 ccRCC 34 M af11N0 MX III 1 + minus + minus
15 Cancer of the renal pelvis 76 M pT3N0M1 III 4 + + + +16 ccRCC 56 F af11N0MX III 1 minus minus + +17 ccRCC 62 F af11N0MX III 1 + minus + +18 ccRCC 46 F af1kN0MX III 1 + + minus minus
19 ccRCC 55 F af2N0MX III 2 + + + minus
20 ccRCC 45 F T2N0M0 II 2 minus + minus minus
21 ccRCC 61 F af11N0MX III 2 + + minus minus
22 ccRCC 60 M NA III 2 + + minus +23 ccRCC 63 F af1kN0MX III 2 + minus minus minus
24 Sarcoma-like 60 F NA III 4 + minus minus minus
25 ccRCC 45 M pT1kN0MX III 2 minus + + +26 ccRCC 63 M af11N0M0 III 1 + + minus +27 ccRCC 73 F af11N0M0 III 1 minus + minus minus
lowastThe results in the Table are presented only for the genes with detected aberrant methylation in cfDNA
Table 4 Diagnostic data analysis for the discrimination of renal cancer patients and healthy subjects using cfDNA methylation of variousgenes alone and in combination
Markers Renal cell carcinoma (119899 = 27) Healthy controls (119899 = 15) 1205942 119901 value Sensitivitylowast Specificitylowastlowast LRRC3B 20 (741) 5 (333) 001 741 667RASSF1 17 (630 ) 1 (67) 00058 629 933FHIT 15 (556) 0 (0) 00003 556 100APC 14 (519) 1 (67) 00034 519 933VHL 0 (0) 0 (0) 0 100ITGA9 0 (0) 0 (0) 0 100RASSF1 or FHIT or APC 25 (923) 2 (133) lt00001 923 867RASSF1 or FHIT 21 (778) 1 (67) lt00001 778 933RASSF1 or APC 21 (778) 1 (67) lt00001 778 933lowastSensitivity was calculated as a percentage of positive results from a number of tested RCC patients lowastlowastspecificity was calculated as a percentage of negativetests from a given number of healthy donors
However LRRC3B showed a low specificity as a markerof cancer since it was methylated in 5 out of 15 (333)healthy donors Methylation of FHIT was not detected in thecfDNA of the control group while methylation of the APCand RASSF1 genes was found in 1 out of 15 (67) healthy
donors Methylation of APC and RASSF1 was detected indifferent healthy individuals (Table 4)
The sensitivity of each of thesemarkers exceeded 50 andwas 519 for APC 63 for RASSF1 and 556 for FHITwhich exhibited the best specificity in our test (100)
6 Disease Markers
Table 5 Receiver-operating characteristic (ROC) curve analyses of cfDNA marker-based models to discriminate between healthy persons(119899 = 15) and renal cancer patients (119899 = 27)lowast
AUC Std error 119901 value 95 LCL 95 UCLConc qPCR 080494 006771 000119 067223 093765Conc qPCR+APC 091852 004205 861E minus 06 08361 100094Conc qPCR+FHIT 091358 004898 110E minus 05 081759 100957Conc qPCR+RASSF1 088148 005986 500E minus 05 076416 099881Conc qPCR+ APC+RASSF1 1 0 106E minus 07 1 1Conc qPCR+APC+FHIT 095802 003018 112E minus 06 089888 101717Conc qPCR+RASSF1A+FHIT 094568 004521 216E minus 06 085708 103428Conc qPCR+APC+FHIT+RASSF1 1 0 106E minus 07 1 1lowastCalculated by binary logistic regression using combination of differentmarkers concentration of cfDNA determined by qPCR (Conc qPCR) andmethylationmarker genes (APC FHIT and RASSF1)
The use of the combined analysis of methylation statusof three genes (RASSF1 FHIT and APC) increased thesensitivity (778ndash923) while the specificity remained high(867ndash933) (Table 4) We did not find any correlationbetween hypermethylation cfDNA concentration and clin-icopathological parameters (grade lymph node metastasisage and sex) in patients with renal cancer
To explore the potential of combined cfDNA concen-tration and gene methylation for RCC diagnostics we per-formed binary logistic regression modelling As a predictorof variables we used cfDNA concentrations measured byquantitative PCR andmethylation ofAPC FHIT andRASSF1genes We built separate models for cfDNA concentrationalone and for all possible combinations of cfDNA concentra-tion and methylation of one two or three genes Predictiveproperties of the models were compared by ROC analysis Asreported above the AUC value for the cfDNA concentrationalone was 08 Addition of one of the genes slightly increasedthe AUC to values 088ndash0918 although these differenceswere not statistically significant as can be seen from 95confidence intervals (Table 5) Addition of two genes ledto further increase of the AUC value up to 1 when usingAPC and RASSF1 Finally the AUC value was 1 when weused the cfDNA concentration and methylation of all threegenes studied The results of ROC analysis are summarisedin Table 5 some of representative ROC curves are shown inFigure 2
4 Discussion
The level of cfDNA in blood plasma could be a universalmarker of malignancy [33] Many studies have shown thatchanges in cfDNA concentration can be correlated withdevelopment prognosis and survival of cancer patients Anincrease of cfDNA concentration was observed in patientswith breast gastric ovary lung colon and prostate cancer[11 34ndash39] It was suggested that an increase of cfDNAconcentration in cancer patients is associated with apoptosisand necrosis of cancer cells in the tumor microenvironment[40] This suggestion was supported by numerous cancer-specific alterations (such as allelic imbalances methylationand mutations) that were found in blood cfDNA (for reviews
00
02
04
06
08
10
Sens
itivi
ty
minus02 1000 02 06 0804
Conc_qPCR
Reference lineConc_qPCR APC+Conc_qPCR APC+ + FHIT
Conc_qPCR APC+ +FHIT + RASSF1A
1minus specificity
Figure 2 Receiver-operating characteristics (ROC) curves obtainedby using different models for the discrimination between healthycontrols (119899 = 15) and renal carcinoma patients (119899 = 27)Conc qPCR concentration of cfDNA determined by qPCR
see [11 41]) It was also demonstrated that monitoring of thecfDNA level in peripheral blood can be used as biomarker ofresponse to therapy in different cancer types [38 42 43]
Previous studies demonstrated that the evaluation ofconcentration of low molecular weight cfDNA (up to 100 bp)is the most representative for detection of malignancies anddisease prognosis since the level of fragments of this sizeincreases with disease progression [44ndash47] Recently Lu etal [47] showed that cfDNA fragments of 67 bp and 180 bpdid not differ between the controls and nonmetastatic RCCpatients while the cfDNA integrity index decreased fromcontrol to the metastatic group Significantly higher concen-trations of low molecular weight fragments were found inthe RCC patients [47] Here we have shown an increase ofcfDNA concentration in RCC patients using genomic cfDNAfragment of 120573-actin gene of 99 bp Recent experiments fromother laboratories also demonstrated increased cfDNA levels
Disease Markers 7
in the blood of patientswith renal cancer compared to healthyindividuals [19 48 49]
Absolute values of cfDNAconcentrations obtained by twodistinct methods are different (11ndash2249 ngmL of plasma inthe fluorescence test compared to 23ndash1177 ngmL in qPCR)but in both cases they were significantly higher than inhealthy individuals (4ndash426 ngmL in the fluorescence testand 3ndash146 ngmL in qPCR) The obtained results agree withdata from other studies in which the determination of thecfDNA concentration using fluorescent dyes gave highervalues than qPCR [24 41] The high AUC values obtainedfor both methods of cfDNA concentration measurement inRCC patients (07679 for fluorescent test and 080494 forqPCR) demonstrate that these methods can be used forclinical investigations In addition to quantitative changescfDNA also possesses qualitative changes that occur in DNAof tumor cells such as mutations microsatellite instabilityand methylation [11 41 50] Methylation of gene promotersis a well-known mechanism of regulation of gene expression[51] Most frequently aberrantmethylation of genes occurs incancer cells Aberrant methylation of the promoter detectedin cfDNA can be used for noninvasive detection of cancerdifferential diagnosis prognosis of survival and response tocancer therapy [52ndash57] Currently several diagnostic systemsbased on the detection of DNA methylation exist that areaimed not only at detecting malignancy (MethylMeter fromRiboMed USA) but also at detecting specific types of cancer(Epi proColon and Epi proLung from Epigenomics AG Ger-many Confirm MDx for Prostate Cancer from MDxHealthUSA Product series DecisionDx G-CIMP Melanom ECUMThymoma from Castle Biosciences) The search for newtools is being pursued because only two of these systems(MethylMeter and Epi proColon) can detect cancer at earlystages of development and can monitor treatment since theyare based on the detection of cfDNA methylation
In this study we started investigating methylation ofpreviously identified tumor suppressor genes in cfDNAData from many studies show that the RASSF1 gene playsan important role in cancerogenesis Hypermethylation ofRASSF1 CpG islands is associated with different types ofcancer and with the risk of progression of tumorigenesis [58ndash64] It was also shown that rat RASSF1 is involved in earlytumorigenesis of RCC [16 17] Studies on methylation of thisgene in blood serum led to controversial results Hauser et al[18] showed that RASSF1A is methylated in 229 of patientsthe study of De Martino et al [19] demonstrated methylationof RASSF1A in 459 of patients Hoque et al [20] observedmethylation of this gene in 11 of serum samples of patientswith RCC In our study methylation of RASSF1 was detectedin 629 of patients The differences in methylation levels ofthe RASSF1 gene can be explained by the use of different CpGislands for analyses We studied methylation of CpG regionlocated within the first exon of RASSF1C while Hauser et al[18] and De Martino et al [19] analysed the region locatedupstream of the initiation codon Previously it was reportedthat these two CpG islands were differentially methylatedin melanoma cell lines and melanoma tumors [65] Ellingeret al [66] demonstrated a 100 correlation between DNAhypermethylation of the RASSF1A promoter and papillary
RCC However De Martino et al [19] analysed 31 samplesof papillary RCCs and found no association of RASSF1Amethylation with the histological subtypes of RCC In ourstudy RASSF1 was also methylated in papillary RCC but itwas the only sample of this cancer subtype analysed
Previously we reported changes in the LRRC3B genepromoter during the search for genetic and epigenetic alter-ations in chromosome 3 in epithelial tumors using NotI-microarrays [14 67 68] LRRC3B was identified by Kim etal [69] as a putative gene suppressor of several tumors thatare silenced in gastric cancers by epigenetic mechanismsIncreased methylation of the LRRC3B gene promoter wasconfirmed in samples of clear cell RCC and colorectalhead and neck cancer [29 70 71] A high level of LRRC3Bhypermethylation was noted not only in RCC patients (74)but also in healthy donors (33) in our study questioning theuse of this gene for the diagnosis of renal cancer on cfDNA
The promoter of the APC gene was methylated in 519of patients which is in good agreement with the results ofHauser and colleagues [18] who detected methylation of theAPC gene in 543 of patients using cfDNA
Previously a significant correlation between FHITexpression in clear cell renal carcinomas and patientsurvival was demonstrated [21] Kvasha et al [22] showeda correlation between hypermethylation of the FHIT CpGisland and a significant decrease of FHIT expression inclear cell RCC The level of aberrant methylation of FHITobtained in our study on cfDNA (556) was close to theresults obtained in the study of Kvasha et al in samples ofRCC tumors (546)
Integrin 12057291205731 plays an important role in various signaltransduction pathways that control proliferation migrationand differentiation of both normal (reviewed in [72 73])and cancer cells (reviewed in [74 75]) Downregulation ofITGA9 expression was observed in several cancer types [76ndash78] that could be caused either by mutations in this gene [79]or by hypermethylation [31 68 80] However methylationof the ITGA9 gene was not detected in our experimentsWe also have not identified methylation of the VHL genealthough NotI-microarray hybridization revealed high levelsof changes in this gene (47) in renal cancer [14] It is possiblethat these changes are associated with deletions in the generather than with methylation At the same time in the studyof DeMartino et al where cfDNAwas analysed by restrictionanalysis methylated VHL was detected in 503 of patientswith RCC [19]
Methylation analysis of the RASSF1 FHIT and APCgenes demonstrated their high specificity (933 for RASSF1and APC 100 for FHIT) for renal tumors Neverthelesssensitivity in one gene analysis was just from 519 forAPC to 629 for RASSF1 (Table 4) At the same timethe use of a combination of three or two genes (withoutLRRC3B due to the low specificity of this gene) leads to asignificant increase in sensitivity (779ndash923) and specificity(867ndash933) All other combinations did not reveal anyadditional diagnosis information Simultaneous methylationof the RASSF1 APC and FHIT genes was identified only in3 patients with metastases However the small sample sizedoes not allow us to draw a conclusion on the correlation
8 Disease Markers
between methylation and disease progression At the sametime binary logistic regression analysis showed the consid-erable diagnostic potential of combining both approachesused in this study According to the ROC analysis the use ofonly cfDNA concentration has moderate diagnostic potential(AUC = 08) On the other hand by using the concentrationand methylation of two or three genes we achieved 100diagnostic accuracy in our samples These results of coursecannot be directly transferred to clinical practice and needverification on a larger number of samples However our datademonstrates the potential advantage there is in combiningevaluation of cfDNA concentration and gene methylation forRCC diagnostics and provides a basis for further research
Thus despite the small sampling our results confirm thepossibility of using the concentration of cfDNA in bloodplasma as an additional marker of renal cancer developmentand show that methylation of three genes FHIT APC andRASSF1 in cfDNA can be used to develop renal cancerdiagnostic tools
5 Conclusion
The results obtained indicate that the concentration of cell-free DNA in plasma and the methylation of specific genes(such as FHIT APC and RASSF1) can be a significantaddition to serological tumor markers in the identification ofpatients with renal cancer However further studies need tobe performed to evaluate their diagnostic value
Competing Interests
The authors declare that they have no competing interests
Acknowledgments
The authors thank Dr A-L Haenni for helpful discussionsand comments on the manuscript This work was supportedby Grants 0110U004534 and 115U002951 from NationalAcademy of Sciences of Ukraine
References
[1] G Banumathy and P Cairns ldquoSignaling pathways in renal cellcarcinomardquo Cancer Biology andTherapy vol 10 no 7 pp 658ndash664 2010
[2] K Gupta J D Miller J Z Li M W Russell and C Charbon-neau ldquoEpidemiologic and socioeconomic burden of metastaticrenal cell carcinoma (mRCC) a literature reviewrdquo CancerTreatment Reviews vol 34 no 3 pp 193ndash205 2008
[3] Ministry of Healthcare of Ukraine The Features Diagnosis andPrognosis Factors Renal Cell Carcinoma Ministry of Healthcareof Ukraine 2011 httpwwwsouuorgua
[4] World Cancer Research Fund InternationalAmerican Institutefor Cancer Research Continuous Update Project Report DietNutrition Physical Activity and Kidney Cancer 2015
[5] A Lopez-Beltran M Scarpelli R Montironi and Z Kirkalildquo2004 WHO classification of the renal tumors of the adultsrdquoEuropean Urology vol 49 no 5 pp 798ndash805 2006
[6] P Hadaczek Z Siprashvili M Markiewski et al ldquoAbsenceor reduction of FHIT expression in most clear cell renalcarcinomasrdquo Cancer Research vol 58 no 14 pp 2946ndash29511998
[7] E Arai and Y Kanai ldquoGenetic and epigenetic alterationsduring renal carcinogenesisrdquo International Journal of Clinicaland Experimental Pathology vol 4 no 1 pp 58ndash73 2011
[8] B N Lasseigne T C Burwell M A Patil D M Absher J DBrooks and R M Myers ldquoDNA methylation profiling revealsnovel diagnostic biomarkers in renal cell carcinomardquo BMCMedicine vol 12 no 1 article 235 2014
[9] N Shenoy N Vallumsetla Y Zou et al ldquoRole of DNAmethylation in renal cell carcinomardquo Journal of Hematology andOncology vol 8 no 1 article 88 2015
[10] G J Fournie J-P Courtin F Laval et al ldquoPlasma DNA asa marker of cancerous cell death Investigations in patientssuffering from lung cancer and in nude mice bearing humantumoursrdquo Cancer Letters vol 91 no 2 pp 221ndash227 1995
[11] H Schwarzenbach D S B Hoon and K Pantel ldquoCell-freenucleic acids as biomarkers in cancer patientsrdquo Nature ReviewsCancer vol 11 no 6 pp 426ndash437 2011
[12] B Taback and D S B Hoon ldquoCirculating nucleic acids andproteomics of plasmaserum clinical utilityrdquo Annals of the NewYork Academy of Sciences vol 1022 pp 1ndash8 2004
[13] R Wasserkort A Kalmar G Valcz et al ldquoAberrant septin 9DNA methylation in colorectal cancer is restricted to a singleCpG islandrdquo BMC Cancer vol 13 article 398 2013
[14] I Ya Skrypkina V I Kashuba V V Gordiyuk et al ldquoIdentifi-cation of changes in gene loci potentially associated with renalcancer by novel technique of NotI microarraysrdquo Reports of theNational Academy of Sciences of Ukraine vol 11 pp 188ndash1922006
[15] V I Kashuba I Y Skrypkina D V Saraev et al ldquoIdentificationof changes in gene loci potentially associated with cervical can-cer usingNotImicroarraysrdquoUkrainrsquoskyi Biokhimichnyi Zhurnalvol 78 no 2 pp 113ndash120 2006
[16] V I Loginov D S Khodyrev I V Pronina et al ldquoMethylationof the RASSF1A promoter region and the allelic imbalancefrequencies in chromosome 3 critical regions correlate withprogression of clear cell renal carcinomardquo Molecular Biologyvol 43 no 3 pp 394ndash402 2009
[17] I Peters K Rehmet N Wilke et al ldquoRASSF1A promotermethylation and expression analysis in normal and neoplastickidney indicates a role in early tumorigenesisrdquo MolecularCancer vol 6 article 49 2007
[18] S Hauser T Zahalka and G Fechner ldquoSerum DNA hyperme-thylation in patients with kidney cancer results of a prospectivestudyrdquo Anticancer Research vol 33 pp 4651ndash4656 2013
[19] M De Martino T Klatte A Haitel and M Marberger ldquoSerumcell-free DNA in renal cell carcinoma a diagnostic and prog-nostic markerrdquo Cancer vol 118 no 1 pp 82ndash90 2012
[20] M O Hoque S Begum O Topaloglu et al ldquoQuantitativedetection of promoter hypermethylation of multiple genes inthe tumor urine and serumDNAof patients with renal cancerrdquoCancer Research vol 64 no 15 pp 5511ndash5517 2004
[21] U Ramp E Caliskan T Ebert et al ldquoFHIT expression inclear cell renal carcinomas versatility of protein levels andcorrelation with survivalrdquoThe Journal of Pathology vol 196 no4 pp 430ndash436 2002
[22] S Kvasha V Gordiyuk A Kondratov et al ldquoHypermethylationof the 51015840CpG island of the FHIT gene in clear cell renalcarcinomasrdquo Cancer Letters vol 265 no 2 pp 250ndash257 2008
Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

6 Disease Markers
Table 5 Receiver-operating characteristic (ROC) curve analyses of cfDNA marker-based models to discriminate between healthy persons(119899 = 15) and renal cancer patients (119899 = 27)lowast
AUC Std error 119901 value 95 LCL 95 UCLConc qPCR 080494 006771 000119 067223 093765Conc qPCR+APC 091852 004205 861E minus 06 08361 100094Conc qPCR+FHIT 091358 004898 110E minus 05 081759 100957Conc qPCR+RASSF1 088148 005986 500E minus 05 076416 099881Conc qPCR+ APC+RASSF1 1 0 106E minus 07 1 1Conc qPCR+APC+FHIT 095802 003018 112E minus 06 089888 101717Conc qPCR+RASSF1A+FHIT 094568 004521 216E minus 06 085708 103428Conc qPCR+APC+FHIT+RASSF1 1 0 106E minus 07 1 1lowastCalculated by binary logistic regression using combination of differentmarkers concentration of cfDNA determined by qPCR (Conc qPCR) andmethylationmarker genes (APC FHIT and RASSF1)
The use of the combined analysis of methylation statusof three genes (RASSF1 FHIT and APC) increased thesensitivity (778ndash923) while the specificity remained high(867ndash933) (Table 4) We did not find any correlationbetween hypermethylation cfDNA concentration and clin-icopathological parameters (grade lymph node metastasisage and sex) in patients with renal cancer
To explore the potential of combined cfDNA concen-tration and gene methylation for RCC diagnostics we per-formed binary logistic regression modelling As a predictorof variables we used cfDNA concentrations measured byquantitative PCR andmethylation ofAPC FHIT andRASSF1genes We built separate models for cfDNA concentrationalone and for all possible combinations of cfDNA concentra-tion and methylation of one two or three genes Predictiveproperties of the models were compared by ROC analysis Asreported above the AUC value for the cfDNA concentrationalone was 08 Addition of one of the genes slightly increasedthe AUC to values 088ndash0918 although these differenceswere not statistically significant as can be seen from 95confidence intervals (Table 5) Addition of two genes ledto further increase of the AUC value up to 1 when usingAPC and RASSF1 Finally the AUC value was 1 when weused the cfDNA concentration and methylation of all threegenes studied The results of ROC analysis are summarisedin Table 5 some of representative ROC curves are shown inFigure 2
4 Discussion
The level of cfDNA in blood plasma could be a universalmarker of malignancy [33] Many studies have shown thatchanges in cfDNA concentration can be correlated withdevelopment prognosis and survival of cancer patients Anincrease of cfDNA concentration was observed in patientswith breast gastric ovary lung colon and prostate cancer[11 34ndash39] It was suggested that an increase of cfDNAconcentration in cancer patients is associated with apoptosisand necrosis of cancer cells in the tumor microenvironment[40] This suggestion was supported by numerous cancer-specific alterations (such as allelic imbalances methylationand mutations) that were found in blood cfDNA (for reviews
00
02
04
06
08
10
Sens
itivi
ty
minus02 1000 02 06 0804
Conc_qPCR
Reference lineConc_qPCR APC+Conc_qPCR APC+ + FHIT
Conc_qPCR APC+ +FHIT + RASSF1A
1minus specificity
Figure 2 Receiver-operating characteristics (ROC) curves obtainedby using different models for the discrimination between healthycontrols (119899 = 15) and renal carcinoma patients (119899 = 27)Conc qPCR concentration of cfDNA determined by qPCR
see [11 41]) It was also demonstrated that monitoring of thecfDNA level in peripheral blood can be used as biomarker ofresponse to therapy in different cancer types [38 42 43]
Previous studies demonstrated that the evaluation ofconcentration of low molecular weight cfDNA (up to 100 bp)is the most representative for detection of malignancies anddisease prognosis since the level of fragments of this sizeincreases with disease progression [44ndash47] Recently Lu etal [47] showed that cfDNA fragments of 67 bp and 180 bpdid not differ between the controls and nonmetastatic RCCpatients while the cfDNA integrity index decreased fromcontrol to the metastatic group Significantly higher concen-trations of low molecular weight fragments were found inthe RCC patients [47] Here we have shown an increase ofcfDNA concentration in RCC patients using genomic cfDNAfragment of 120573-actin gene of 99 bp Recent experiments fromother laboratories also demonstrated increased cfDNA levels
Disease Markers 7
in the blood of patientswith renal cancer compared to healthyindividuals [19 48 49]
Absolute values of cfDNAconcentrations obtained by twodistinct methods are different (11ndash2249 ngmL of plasma inthe fluorescence test compared to 23ndash1177 ngmL in qPCR)but in both cases they were significantly higher than inhealthy individuals (4ndash426 ngmL in the fluorescence testand 3ndash146 ngmL in qPCR) The obtained results agree withdata from other studies in which the determination of thecfDNA concentration using fluorescent dyes gave highervalues than qPCR [24 41] The high AUC values obtainedfor both methods of cfDNA concentration measurement inRCC patients (07679 for fluorescent test and 080494 forqPCR) demonstrate that these methods can be used forclinical investigations In addition to quantitative changescfDNA also possesses qualitative changes that occur in DNAof tumor cells such as mutations microsatellite instabilityand methylation [11 41 50] Methylation of gene promotersis a well-known mechanism of regulation of gene expression[51] Most frequently aberrantmethylation of genes occurs incancer cells Aberrant methylation of the promoter detectedin cfDNA can be used for noninvasive detection of cancerdifferential diagnosis prognosis of survival and response tocancer therapy [52ndash57] Currently several diagnostic systemsbased on the detection of DNA methylation exist that areaimed not only at detecting malignancy (MethylMeter fromRiboMed USA) but also at detecting specific types of cancer(Epi proColon and Epi proLung from Epigenomics AG Ger-many Confirm MDx for Prostate Cancer from MDxHealthUSA Product series DecisionDx G-CIMP Melanom ECUMThymoma from Castle Biosciences) The search for newtools is being pursued because only two of these systems(MethylMeter and Epi proColon) can detect cancer at earlystages of development and can monitor treatment since theyare based on the detection of cfDNA methylation
In this study we started investigating methylation ofpreviously identified tumor suppressor genes in cfDNAData from many studies show that the RASSF1 gene playsan important role in cancerogenesis Hypermethylation ofRASSF1 CpG islands is associated with different types ofcancer and with the risk of progression of tumorigenesis [58ndash64] It was also shown that rat RASSF1 is involved in earlytumorigenesis of RCC [16 17] Studies on methylation of thisgene in blood serum led to controversial results Hauser et al[18] showed that RASSF1A is methylated in 229 of patientsthe study of De Martino et al [19] demonstrated methylationof RASSF1A in 459 of patients Hoque et al [20] observedmethylation of this gene in 11 of serum samples of patientswith RCC In our study methylation of RASSF1 was detectedin 629 of patients The differences in methylation levels ofthe RASSF1 gene can be explained by the use of different CpGislands for analyses We studied methylation of CpG regionlocated within the first exon of RASSF1C while Hauser et al[18] and De Martino et al [19] analysed the region locatedupstream of the initiation codon Previously it was reportedthat these two CpG islands were differentially methylatedin melanoma cell lines and melanoma tumors [65] Ellingeret al [66] demonstrated a 100 correlation between DNAhypermethylation of the RASSF1A promoter and papillary
RCC However De Martino et al [19] analysed 31 samplesof papillary RCCs and found no association of RASSF1Amethylation with the histological subtypes of RCC In ourstudy RASSF1 was also methylated in papillary RCC but itwas the only sample of this cancer subtype analysed
Previously we reported changes in the LRRC3B genepromoter during the search for genetic and epigenetic alter-ations in chromosome 3 in epithelial tumors using NotI-microarrays [14 67 68] LRRC3B was identified by Kim etal [69] as a putative gene suppressor of several tumors thatare silenced in gastric cancers by epigenetic mechanismsIncreased methylation of the LRRC3B gene promoter wasconfirmed in samples of clear cell RCC and colorectalhead and neck cancer [29 70 71] A high level of LRRC3Bhypermethylation was noted not only in RCC patients (74)but also in healthy donors (33) in our study questioning theuse of this gene for the diagnosis of renal cancer on cfDNA
The promoter of the APC gene was methylated in 519of patients which is in good agreement with the results ofHauser and colleagues [18] who detected methylation of theAPC gene in 543 of patients using cfDNA
Previously a significant correlation between FHITexpression in clear cell renal carcinomas and patientsurvival was demonstrated [21] Kvasha et al [22] showeda correlation between hypermethylation of the FHIT CpGisland and a significant decrease of FHIT expression inclear cell RCC The level of aberrant methylation of FHITobtained in our study on cfDNA (556) was close to theresults obtained in the study of Kvasha et al in samples ofRCC tumors (546)
Integrin 12057291205731 plays an important role in various signaltransduction pathways that control proliferation migrationand differentiation of both normal (reviewed in [72 73])and cancer cells (reviewed in [74 75]) Downregulation ofITGA9 expression was observed in several cancer types [76ndash78] that could be caused either by mutations in this gene [79]or by hypermethylation [31 68 80] However methylationof the ITGA9 gene was not detected in our experimentsWe also have not identified methylation of the VHL genealthough NotI-microarray hybridization revealed high levelsof changes in this gene (47) in renal cancer [14] It is possiblethat these changes are associated with deletions in the generather than with methylation At the same time in the studyof DeMartino et al where cfDNAwas analysed by restrictionanalysis methylated VHL was detected in 503 of patientswith RCC [19]
Methylation analysis of the RASSF1 FHIT and APCgenes demonstrated their high specificity (933 for RASSF1and APC 100 for FHIT) for renal tumors Neverthelesssensitivity in one gene analysis was just from 519 forAPC to 629 for RASSF1 (Table 4) At the same timethe use of a combination of three or two genes (withoutLRRC3B due to the low specificity of this gene) leads to asignificant increase in sensitivity (779ndash923) and specificity(867ndash933) All other combinations did not reveal anyadditional diagnosis information Simultaneous methylationof the RASSF1 APC and FHIT genes was identified only in3 patients with metastases However the small sample sizedoes not allow us to draw a conclusion on the correlation
8 Disease Markers
between methylation and disease progression At the sametime binary logistic regression analysis showed the consid-erable diagnostic potential of combining both approachesused in this study According to the ROC analysis the use ofonly cfDNA concentration has moderate diagnostic potential(AUC = 08) On the other hand by using the concentrationand methylation of two or three genes we achieved 100diagnostic accuracy in our samples These results of coursecannot be directly transferred to clinical practice and needverification on a larger number of samples However our datademonstrates the potential advantage there is in combiningevaluation of cfDNA concentration and gene methylation forRCC diagnostics and provides a basis for further research
Thus despite the small sampling our results confirm thepossibility of using the concentration of cfDNA in bloodplasma as an additional marker of renal cancer developmentand show that methylation of three genes FHIT APC andRASSF1 in cfDNA can be used to develop renal cancerdiagnostic tools
5 Conclusion
The results obtained indicate that the concentration of cell-free DNA in plasma and the methylation of specific genes(such as FHIT APC and RASSF1) can be a significantaddition to serological tumor markers in the identification ofpatients with renal cancer However further studies need tobe performed to evaluate their diagnostic value
Competing Interests
The authors declare that they have no competing interests
Acknowledgments
The authors thank Dr A-L Haenni for helpful discussionsand comments on the manuscript This work was supportedby Grants 0110U004534 and 115U002951 from NationalAcademy of Sciences of Ukraine
References
[1] G Banumathy and P Cairns ldquoSignaling pathways in renal cellcarcinomardquo Cancer Biology andTherapy vol 10 no 7 pp 658ndash664 2010
[2] K Gupta J D Miller J Z Li M W Russell and C Charbon-neau ldquoEpidemiologic and socioeconomic burden of metastaticrenal cell carcinoma (mRCC) a literature reviewrdquo CancerTreatment Reviews vol 34 no 3 pp 193ndash205 2008
[3] Ministry of Healthcare of Ukraine The Features Diagnosis andPrognosis Factors Renal Cell Carcinoma Ministry of Healthcareof Ukraine 2011 httpwwwsouuorgua
[4] World Cancer Research Fund InternationalAmerican Institutefor Cancer Research Continuous Update Project Report DietNutrition Physical Activity and Kidney Cancer 2015
[5] A Lopez-Beltran M Scarpelli R Montironi and Z Kirkalildquo2004 WHO classification of the renal tumors of the adultsrdquoEuropean Urology vol 49 no 5 pp 798ndash805 2006
[6] P Hadaczek Z Siprashvili M Markiewski et al ldquoAbsenceor reduction of FHIT expression in most clear cell renalcarcinomasrdquo Cancer Research vol 58 no 14 pp 2946ndash29511998
[7] E Arai and Y Kanai ldquoGenetic and epigenetic alterationsduring renal carcinogenesisrdquo International Journal of Clinicaland Experimental Pathology vol 4 no 1 pp 58ndash73 2011
[8] B N Lasseigne T C Burwell M A Patil D M Absher J DBrooks and R M Myers ldquoDNA methylation profiling revealsnovel diagnostic biomarkers in renal cell carcinomardquo BMCMedicine vol 12 no 1 article 235 2014
[9] N Shenoy N Vallumsetla Y Zou et al ldquoRole of DNAmethylation in renal cell carcinomardquo Journal of Hematology andOncology vol 8 no 1 article 88 2015
[10] G J Fournie J-P Courtin F Laval et al ldquoPlasma DNA asa marker of cancerous cell death Investigations in patientssuffering from lung cancer and in nude mice bearing humantumoursrdquo Cancer Letters vol 91 no 2 pp 221ndash227 1995
[11] H Schwarzenbach D S B Hoon and K Pantel ldquoCell-freenucleic acids as biomarkers in cancer patientsrdquo Nature ReviewsCancer vol 11 no 6 pp 426ndash437 2011
[12] B Taback and D S B Hoon ldquoCirculating nucleic acids andproteomics of plasmaserum clinical utilityrdquo Annals of the NewYork Academy of Sciences vol 1022 pp 1ndash8 2004
[13] R Wasserkort A Kalmar G Valcz et al ldquoAberrant septin 9DNA methylation in colorectal cancer is restricted to a singleCpG islandrdquo BMC Cancer vol 13 article 398 2013
[14] I Ya Skrypkina V I Kashuba V V Gordiyuk et al ldquoIdentifi-cation of changes in gene loci potentially associated with renalcancer by novel technique of NotI microarraysrdquo Reports of theNational Academy of Sciences of Ukraine vol 11 pp 188ndash1922006
[15] V I Kashuba I Y Skrypkina D V Saraev et al ldquoIdentificationof changes in gene loci potentially associated with cervical can-cer usingNotImicroarraysrdquoUkrainrsquoskyi Biokhimichnyi Zhurnalvol 78 no 2 pp 113ndash120 2006
[16] V I Loginov D S Khodyrev I V Pronina et al ldquoMethylationof the RASSF1A promoter region and the allelic imbalancefrequencies in chromosome 3 critical regions correlate withprogression of clear cell renal carcinomardquo Molecular Biologyvol 43 no 3 pp 394ndash402 2009
[17] I Peters K Rehmet N Wilke et al ldquoRASSF1A promotermethylation and expression analysis in normal and neoplastickidney indicates a role in early tumorigenesisrdquo MolecularCancer vol 6 article 49 2007
[18] S Hauser T Zahalka and G Fechner ldquoSerum DNA hyperme-thylation in patients with kidney cancer results of a prospectivestudyrdquo Anticancer Research vol 33 pp 4651ndash4656 2013
[19] M De Martino T Klatte A Haitel and M Marberger ldquoSerumcell-free DNA in renal cell carcinoma a diagnostic and prog-nostic markerrdquo Cancer vol 118 no 1 pp 82ndash90 2012
[20] M O Hoque S Begum O Topaloglu et al ldquoQuantitativedetection of promoter hypermethylation of multiple genes inthe tumor urine and serumDNAof patients with renal cancerrdquoCancer Research vol 64 no 15 pp 5511ndash5517 2004
[21] U Ramp E Caliskan T Ebert et al ldquoFHIT expression inclear cell renal carcinomas versatility of protein levels andcorrelation with survivalrdquoThe Journal of Pathology vol 196 no4 pp 430ndash436 2002
[22] S Kvasha V Gordiyuk A Kondratov et al ldquoHypermethylationof the 51015840CpG island of the FHIT gene in clear cell renalcarcinomasrdquo Cancer Letters vol 265 no 2 pp 250ndash257 2008
Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

Disease Markers 7
in the blood of patientswith renal cancer compared to healthyindividuals [19 48 49]
Absolute values of cfDNAconcentrations obtained by twodistinct methods are different (11ndash2249 ngmL of plasma inthe fluorescence test compared to 23ndash1177 ngmL in qPCR)but in both cases they were significantly higher than inhealthy individuals (4ndash426 ngmL in the fluorescence testand 3ndash146 ngmL in qPCR) The obtained results agree withdata from other studies in which the determination of thecfDNA concentration using fluorescent dyes gave highervalues than qPCR [24 41] The high AUC values obtainedfor both methods of cfDNA concentration measurement inRCC patients (07679 for fluorescent test and 080494 forqPCR) demonstrate that these methods can be used forclinical investigations In addition to quantitative changescfDNA also possesses qualitative changes that occur in DNAof tumor cells such as mutations microsatellite instabilityand methylation [11 41 50] Methylation of gene promotersis a well-known mechanism of regulation of gene expression[51] Most frequently aberrantmethylation of genes occurs incancer cells Aberrant methylation of the promoter detectedin cfDNA can be used for noninvasive detection of cancerdifferential diagnosis prognosis of survival and response tocancer therapy [52ndash57] Currently several diagnostic systemsbased on the detection of DNA methylation exist that areaimed not only at detecting malignancy (MethylMeter fromRiboMed USA) but also at detecting specific types of cancer(Epi proColon and Epi proLung from Epigenomics AG Ger-many Confirm MDx for Prostate Cancer from MDxHealthUSA Product series DecisionDx G-CIMP Melanom ECUMThymoma from Castle Biosciences) The search for newtools is being pursued because only two of these systems(MethylMeter and Epi proColon) can detect cancer at earlystages of development and can monitor treatment since theyare based on the detection of cfDNA methylation
In this study we started investigating methylation ofpreviously identified tumor suppressor genes in cfDNAData from many studies show that the RASSF1 gene playsan important role in cancerogenesis Hypermethylation ofRASSF1 CpG islands is associated with different types ofcancer and with the risk of progression of tumorigenesis [58ndash64] It was also shown that rat RASSF1 is involved in earlytumorigenesis of RCC [16 17] Studies on methylation of thisgene in blood serum led to controversial results Hauser et al[18] showed that RASSF1A is methylated in 229 of patientsthe study of De Martino et al [19] demonstrated methylationof RASSF1A in 459 of patients Hoque et al [20] observedmethylation of this gene in 11 of serum samples of patientswith RCC In our study methylation of RASSF1 was detectedin 629 of patients The differences in methylation levels ofthe RASSF1 gene can be explained by the use of different CpGislands for analyses We studied methylation of CpG regionlocated within the first exon of RASSF1C while Hauser et al[18] and De Martino et al [19] analysed the region locatedupstream of the initiation codon Previously it was reportedthat these two CpG islands were differentially methylatedin melanoma cell lines and melanoma tumors [65] Ellingeret al [66] demonstrated a 100 correlation between DNAhypermethylation of the RASSF1A promoter and papillary
RCC However De Martino et al [19] analysed 31 samplesof papillary RCCs and found no association of RASSF1Amethylation with the histological subtypes of RCC In ourstudy RASSF1 was also methylated in papillary RCC but itwas the only sample of this cancer subtype analysed
Previously we reported changes in the LRRC3B genepromoter during the search for genetic and epigenetic alter-ations in chromosome 3 in epithelial tumors using NotI-microarrays [14 67 68] LRRC3B was identified by Kim etal [69] as a putative gene suppressor of several tumors thatare silenced in gastric cancers by epigenetic mechanismsIncreased methylation of the LRRC3B gene promoter wasconfirmed in samples of clear cell RCC and colorectalhead and neck cancer [29 70 71] A high level of LRRC3Bhypermethylation was noted not only in RCC patients (74)but also in healthy donors (33) in our study questioning theuse of this gene for the diagnosis of renal cancer on cfDNA
The promoter of the APC gene was methylated in 519of patients which is in good agreement with the results ofHauser and colleagues [18] who detected methylation of theAPC gene in 543 of patients using cfDNA
Previously a significant correlation between FHITexpression in clear cell renal carcinomas and patientsurvival was demonstrated [21] Kvasha et al [22] showeda correlation between hypermethylation of the FHIT CpGisland and a significant decrease of FHIT expression inclear cell RCC The level of aberrant methylation of FHITobtained in our study on cfDNA (556) was close to theresults obtained in the study of Kvasha et al in samples ofRCC tumors (546)
Integrin 12057291205731 plays an important role in various signaltransduction pathways that control proliferation migrationand differentiation of both normal (reviewed in [72 73])and cancer cells (reviewed in [74 75]) Downregulation ofITGA9 expression was observed in several cancer types [76ndash78] that could be caused either by mutations in this gene [79]or by hypermethylation [31 68 80] However methylationof the ITGA9 gene was not detected in our experimentsWe also have not identified methylation of the VHL genealthough NotI-microarray hybridization revealed high levelsof changes in this gene (47) in renal cancer [14] It is possiblethat these changes are associated with deletions in the generather than with methylation At the same time in the studyof DeMartino et al where cfDNAwas analysed by restrictionanalysis methylated VHL was detected in 503 of patientswith RCC [19]
Methylation analysis of the RASSF1 FHIT and APCgenes demonstrated their high specificity (933 for RASSF1and APC 100 for FHIT) for renal tumors Neverthelesssensitivity in one gene analysis was just from 519 forAPC to 629 for RASSF1 (Table 4) At the same timethe use of a combination of three or two genes (withoutLRRC3B due to the low specificity of this gene) leads to asignificant increase in sensitivity (779ndash923) and specificity(867ndash933) All other combinations did not reveal anyadditional diagnosis information Simultaneous methylationof the RASSF1 APC and FHIT genes was identified only in3 patients with metastases However the small sample sizedoes not allow us to draw a conclusion on the correlation
8 Disease Markers
between methylation and disease progression At the sametime binary logistic regression analysis showed the consid-erable diagnostic potential of combining both approachesused in this study According to the ROC analysis the use ofonly cfDNA concentration has moderate diagnostic potential(AUC = 08) On the other hand by using the concentrationand methylation of two or three genes we achieved 100diagnostic accuracy in our samples These results of coursecannot be directly transferred to clinical practice and needverification on a larger number of samples However our datademonstrates the potential advantage there is in combiningevaluation of cfDNA concentration and gene methylation forRCC diagnostics and provides a basis for further research
Thus despite the small sampling our results confirm thepossibility of using the concentration of cfDNA in bloodplasma as an additional marker of renal cancer developmentand show that methylation of three genes FHIT APC andRASSF1 in cfDNA can be used to develop renal cancerdiagnostic tools
5 Conclusion
The results obtained indicate that the concentration of cell-free DNA in plasma and the methylation of specific genes(such as FHIT APC and RASSF1) can be a significantaddition to serological tumor markers in the identification ofpatients with renal cancer However further studies need tobe performed to evaluate their diagnostic value
Competing Interests
The authors declare that they have no competing interests
Acknowledgments
The authors thank Dr A-L Haenni for helpful discussionsand comments on the manuscript This work was supportedby Grants 0110U004534 and 115U002951 from NationalAcademy of Sciences of Ukraine
References
[1] G Banumathy and P Cairns ldquoSignaling pathways in renal cellcarcinomardquo Cancer Biology andTherapy vol 10 no 7 pp 658ndash664 2010
[2] K Gupta J D Miller J Z Li M W Russell and C Charbon-neau ldquoEpidemiologic and socioeconomic burden of metastaticrenal cell carcinoma (mRCC) a literature reviewrdquo CancerTreatment Reviews vol 34 no 3 pp 193ndash205 2008
[3] Ministry of Healthcare of Ukraine The Features Diagnosis andPrognosis Factors Renal Cell Carcinoma Ministry of Healthcareof Ukraine 2011 httpwwwsouuorgua
[4] World Cancer Research Fund InternationalAmerican Institutefor Cancer Research Continuous Update Project Report DietNutrition Physical Activity and Kidney Cancer 2015
[5] A Lopez-Beltran M Scarpelli R Montironi and Z Kirkalildquo2004 WHO classification of the renal tumors of the adultsrdquoEuropean Urology vol 49 no 5 pp 798ndash805 2006
[6] P Hadaczek Z Siprashvili M Markiewski et al ldquoAbsenceor reduction of FHIT expression in most clear cell renalcarcinomasrdquo Cancer Research vol 58 no 14 pp 2946ndash29511998
[7] E Arai and Y Kanai ldquoGenetic and epigenetic alterationsduring renal carcinogenesisrdquo International Journal of Clinicaland Experimental Pathology vol 4 no 1 pp 58ndash73 2011
[8] B N Lasseigne T C Burwell M A Patil D M Absher J DBrooks and R M Myers ldquoDNA methylation profiling revealsnovel diagnostic biomarkers in renal cell carcinomardquo BMCMedicine vol 12 no 1 article 235 2014
[9] N Shenoy N Vallumsetla Y Zou et al ldquoRole of DNAmethylation in renal cell carcinomardquo Journal of Hematology andOncology vol 8 no 1 article 88 2015
[10] G J Fournie J-P Courtin F Laval et al ldquoPlasma DNA asa marker of cancerous cell death Investigations in patientssuffering from lung cancer and in nude mice bearing humantumoursrdquo Cancer Letters vol 91 no 2 pp 221ndash227 1995
[11] H Schwarzenbach D S B Hoon and K Pantel ldquoCell-freenucleic acids as biomarkers in cancer patientsrdquo Nature ReviewsCancer vol 11 no 6 pp 426ndash437 2011
[12] B Taback and D S B Hoon ldquoCirculating nucleic acids andproteomics of plasmaserum clinical utilityrdquo Annals of the NewYork Academy of Sciences vol 1022 pp 1ndash8 2004
[13] R Wasserkort A Kalmar G Valcz et al ldquoAberrant septin 9DNA methylation in colorectal cancer is restricted to a singleCpG islandrdquo BMC Cancer vol 13 article 398 2013
[14] I Ya Skrypkina V I Kashuba V V Gordiyuk et al ldquoIdentifi-cation of changes in gene loci potentially associated with renalcancer by novel technique of NotI microarraysrdquo Reports of theNational Academy of Sciences of Ukraine vol 11 pp 188ndash1922006
[15] V I Kashuba I Y Skrypkina D V Saraev et al ldquoIdentificationof changes in gene loci potentially associated with cervical can-cer usingNotImicroarraysrdquoUkrainrsquoskyi Biokhimichnyi Zhurnalvol 78 no 2 pp 113ndash120 2006
[16] V I Loginov D S Khodyrev I V Pronina et al ldquoMethylationof the RASSF1A promoter region and the allelic imbalancefrequencies in chromosome 3 critical regions correlate withprogression of clear cell renal carcinomardquo Molecular Biologyvol 43 no 3 pp 394ndash402 2009
[17] I Peters K Rehmet N Wilke et al ldquoRASSF1A promotermethylation and expression analysis in normal and neoplastickidney indicates a role in early tumorigenesisrdquo MolecularCancer vol 6 article 49 2007
[18] S Hauser T Zahalka and G Fechner ldquoSerum DNA hyperme-thylation in patients with kidney cancer results of a prospectivestudyrdquo Anticancer Research vol 33 pp 4651ndash4656 2013
[19] M De Martino T Klatte A Haitel and M Marberger ldquoSerumcell-free DNA in renal cell carcinoma a diagnostic and prog-nostic markerrdquo Cancer vol 118 no 1 pp 82ndash90 2012
[20] M O Hoque S Begum O Topaloglu et al ldquoQuantitativedetection of promoter hypermethylation of multiple genes inthe tumor urine and serumDNAof patients with renal cancerrdquoCancer Research vol 64 no 15 pp 5511ndash5517 2004
[21] U Ramp E Caliskan T Ebert et al ldquoFHIT expression inclear cell renal carcinomas versatility of protein levels andcorrelation with survivalrdquoThe Journal of Pathology vol 196 no4 pp 430ndash436 2002
[22] S Kvasha V Gordiyuk A Kondratov et al ldquoHypermethylationof the 51015840CpG island of the FHIT gene in clear cell renalcarcinomasrdquo Cancer Letters vol 265 no 2 pp 250ndash257 2008
Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

8 Disease Markers
between methylation and disease progression At the sametime binary logistic regression analysis showed the consid-erable diagnostic potential of combining both approachesused in this study According to the ROC analysis the use ofonly cfDNA concentration has moderate diagnostic potential(AUC = 08) On the other hand by using the concentrationand methylation of two or three genes we achieved 100diagnostic accuracy in our samples These results of coursecannot be directly transferred to clinical practice and needverification on a larger number of samples However our datademonstrates the potential advantage there is in combiningevaluation of cfDNA concentration and gene methylation forRCC diagnostics and provides a basis for further research
Thus despite the small sampling our results confirm thepossibility of using the concentration of cfDNA in bloodplasma as an additional marker of renal cancer developmentand show that methylation of three genes FHIT APC andRASSF1 in cfDNA can be used to develop renal cancerdiagnostic tools
5 Conclusion
The results obtained indicate that the concentration of cell-free DNA in plasma and the methylation of specific genes(such as FHIT APC and RASSF1) can be a significantaddition to serological tumor markers in the identification ofpatients with renal cancer However further studies need tobe performed to evaluate their diagnostic value
Competing Interests
The authors declare that they have no competing interests
Acknowledgments
The authors thank Dr A-L Haenni for helpful discussionsand comments on the manuscript This work was supportedby Grants 0110U004534 and 115U002951 from NationalAcademy of Sciences of Ukraine
References
[1] G Banumathy and P Cairns ldquoSignaling pathways in renal cellcarcinomardquo Cancer Biology andTherapy vol 10 no 7 pp 658ndash664 2010
[2] K Gupta J D Miller J Z Li M W Russell and C Charbon-neau ldquoEpidemiologic and socioeconomic burden of metastaticrenal cell carcinoma (mRCC) a literature reviewrdquo CancerTreatment Reviews vol 34 no 3 pp 193ndash205 2008
[3] Ministry of Healthcare of Ukraine The Features Diagnosis andPrognosis Factors Renal Cell Carcinoma Ministry of Healthcareof Ukraine 2011 httpwwwsouuorgua
[4] World Cancer Research Fund InternationalAmerican Institutefor Cancer Research Continuous Update Project Report DietNutrition Physical Activity and Kidney Cancer 2015
[5] A Lopez-Beltran M Scarpelli R Montironi and Z Kirkalildquo2004 WHO classification of the renal tumors of the adultsrdquoEuropean Urology vol 49 no 5 pp 798ndash805 2006
[6] P Hadaczek Z Siprashvili M Markiewski et al ldquoAbsenceor reduction of FHIT expression in most clear cell renalcarcinomasrdquo Cancer Research vol 58 no 14 pp 2946ndash29511998
[7] E Arai and Y Kanai ldquoGenetic and epigenetic alterationsduring renal carcinogenesisrdquo International Journal of Clinicaland Experimental Pathology vol 4 no 1 pp 58ndash73 2011
[8] B N Lasseigne T C Burwell M A Patil D M Absher J DBrooks and R M Myers ldquoDNA methylation profiling revealsnovel diagnostic biomarkers in renal cell carcinomardquo BMCMedicine vol 12 no 1 article 235 2014
[9] N Shenoy N Vallumsetla Y Zou et al ldquoRole of DNAmethylation in renal cell carcinomardquo Journal of Hematology andOncology vol 8 no 1 article 88 2015
[10] G J Fournie J-P Courtin F Laval et al ldquoPlasma DNA asa marker of cancerous cell death Investigations in patientssuffering from lung cancer and in nude mice bearing humantumoursrdquo Cancer Letters vol 91 no 2 pp 221ndash227 1995
[11] H Schwarzenbach D S B Hoon and K Pantel ldquoCell-freenucleic acids as biomarkers in cancer patientsrdquo Nature ReviewsCancer vol 11 no 6 pp 426ndash437 2011
[12] B Taback and D S B Hoon ldquoCirculating nucleic acids andproteomics of plasmaserum clinical utilityrdquo Annals of the NewYork Academy of Sciences vol 1022 pp 1ndash8 2004
[13] R Wasserkort A Kalmar G Valcz et al ldquoAberrant septin 9DNA methylation in colorectal cancer is restricted to a singleCpG islandrdquo BMC Cancer vol 13 article 398 2013
[14] I Ya Skrypkina V I Kashuba V V Gordiyuk et al ldquoIdentifi-cation of changes in gene loci potentially associated with renalcancer by novel technique of NotI microarraysrdquo Reports of theNational Academy of Sciences of Ukraine vol 11 pp 188ndash1922006
[15] V I Kashuba I Y Skrypkina D V Saraev et al ldquoIdentificationof changes in gene loci potentially associated with cervical can-cer usingNotImicroarraysrdquoUkrainrsquoskyi Biokhimichnyi Zhurnalvol 78 no 2 pp 113ndash120 2006
[16] V I Loginov D S Khodyrev I V Pronina et al ldquoMethylationof the RASSF1A promoter region and the allelic imbalancefrequencies in chromosome 3 critical regions correlate withprogression of clear cell renal carcinomardquo Molecular Biologyvol 43 no 3 pp 394ndash402 2009
[17] I Peters K Rehmet N Wilke et al ldquoRASSF1A promotermethylation and expression analysis in normal and neoplastickidney indicates a role in early tumorigenesisrdquo MolecularCancer vol 6 article 49 2007
[18] S Hauser T Zahalka and G Fechner ldquoSerum DNA hyperme-thylation in patients with kidney cancer results of a prospectivestudyrdquo Anticancer Research vol 33 pp 4651ndash4656 2013
[19] M De Martino T Klatte A Haitel and M Marberger ldquoSerumcell-free DNA in renal cell carcinoma a diagnostic and prog-nostic markerrdquo Cancer vol 118 no 1 pp 82ndash90 2012
[20] M O Hoque S Begum O Topaloglu et al ldquoQuantitativedetection of promoter hypermethylation of multiple genes inthe tumor urine and serumDNAof patients with renal cancerrdquoCancer Research vol 64 no 15 pp 5511ndash5517 2004
[21] U Ramp E Caliskan T Ebert et al ldquoFHIT expression inclear cell renal carcinomas versatility of protein levels andcorrelation with survivalrdquoThe Journal of Pathology vol 196 no4 pp 430ndash436 2002
[22] S Kvasha V Gordiyuk A Kondratov et al ldquoHypermethylationof the 51015840CpG island of the FHIT gene in clear cell renalcarcinomasrdquo Cancer Letters vol 265 no 2 pp 250ndash257 2008
Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

Disease Markers 9
[23] L J Herrera S RajaW E Gooding et al ldquoQuantitative analysisof circulating plasma DNA as a tumor marker in thoracicmalignanciesrdquo Clinical Chemistry vol 51 no 1 pp 113ndash1182005
[24] D Czeiger G Shaked H Eini et al ldquoMeasurement of circu-lating cell-free DNA levels by a new simple fluorescent test inpatients with primary colorectal cancerrdquo American Journal ofClinical Pathology vol 135 no 2 pp 264ndash270 2011
[25] J G Herman J R Graff S Myohanen B D Nelkin and SB Baylin ldquoMethylation-specific PCR a novel PCR assay formethylation status of CpG islandsrdquo Proceedings of the NationalAcademy of Sciences of the United States of America vol 93 no18 pp 9821ndash9826 1996
[26] K Lo J Kwong A B Hui et al ldquoAdvances in brief high fre-quency of promoter hypermethylation of RASSF1A in nasopha-ryngeal carcinomardquo Journal of Clinical Oncology pp 3877ndash38812001
[27] S Zochbauer-Muller K M Fong A Maitra et al ldquo51015840 CpGisland methylation of the FHIT gene is correlated with loss ofgene expression in lung and breast cancerrdquoCancer Research vol61 no 9 pp 3581ndash3585 2001
[28] M Shinozaki D S B Hoon A E Giuliano et al ldquoDistincthypermethylation profile of primary breast cancer is associatedwith sentinel lymph node metastasisrdquo Clinical Cancer Researchvol 11 no 6 pp 2156ndash2162 2005
[29] A G Kondratov L A Stoliar S M Kvasha et al ldquoMethylationpattern of the putative tumor-suppressor gene LRRC3B pro-moter in clear cell renal cell carcinomasrdquo Molecular MedicineReports vol 5 no 2 pp 509ndash512 2012
[30] T Kuroki F Trapasso S Yendamuri et al ldquoAllele loss andpromoter hypermethylation of VHL RAR-120573 RASSF1A andFHIT tumor suppressor genes on chromosome 3p in esophagealsquamous cell carcinomardquo Cancer Research vol 63 no 13 pp3724ndash3728 2003
[31] J-L Li Q Fei J Yu H-Y Zhang PWang and J De Zhu ldquoCor-relation between methylation profile of promoter cpg islands ofseven metastasis-associated genes and their expression states insix cell lines of liver originrdquoAi Zheng vol 23 no 9 pp 985ndash9912004
[32] S Bhalerao and P Kadam ldquoSample size calculationrdquo Interna-tional Journal of Ayurveda Research vol 1 no 1 pp 55ndash57 2010
[33] E Heitzer P Ulz and J B Geigl ldquoCirculating tumor DNA as aliquid biopsy for cancerrdquo Clinical Chemistry vol 61 no 1 pp112ndash123 2015
[34] C Roth K Pantel V Muller et al ldquoApoptosis-related deregula-tion of proteolytic activities and high serum levels of circulatingnucleosomes and DNA in blood correlate with breast cancerprogressionrdquo BMC Cancer vol 11 no 1 article 4 2011
[35] S Sai D Ichikawa H Tomita et al ldquoQuantification of plasmacell-free DNA in patients with gastric cancerrdquo AnticancerResearch vol 27 no 4 pp 2747ndash2751 2007
[36] T-L Wu D Zhang J-H Chia K-C Tsao C-F Sun and J TWu ldquoCell-free DNA measurement in various carcinomas andestablishment of normal reference rangerdquoClinica Chimica Actavol 321 no 1-2 pp 77ndash87 2002
[37] C Bettegowda M Sausen R J Leary et al ldquoDetection ofcirculating tumor DNA in early- and late-stage human malig-nanciesrdquo Science Translational Medicine vol 6 no 224 ArticleID 224ra24 2014
[38] G Feng X Ye F Fang C Pu H Huang and G Li ldquoQuantifica-tion of plasma cell-free DNA in predicting therapeutic efficacy
of sorafenib on metastatic clear cell renal cell carcinomardquoDisease Markers vol 34 no 2 pp 105ndash111 2013
[39] A Kienel D Porres A Heidenreich and D Pfister ldquocfDNA asa prognostic marker of response to taxane based chemotherapyin patients with prostate cancerrdquoThe Journal of Urology vol 194no 4 pp 966ndash971 2015
[40] Z Chen A Fadiel F Naftolin K D Eichenbaum and Y XialdquoCirculationDNA biological implications for cancermetastasisand immunologyrdquo Medical Hypotheses vol 65 no 5 pp 956ndash961 2005
[41] K Jung M Fleischhacker and A Rabien ldquoCell-free DNA inthe blood as a solid tumor biomarkermdasha critical appraisal ofthe literaturerdquo Clinica Chimica Acta vol 411 no 21-22 pp 1611ndash1624 2010
[42] A A Kamat F Z Bischoff D Dang et al ldquoCirculating cell-free DNA a novel biomarker for response to therapy in ovariancarcinomardquoCancer Biology andTherapy vol 5 no 10 pp 1369ndash1374 2006
[43] C Cheng M Omura-Minamisawa Y Kang T Hara I Koikeand T Inoue ldquoQuantification of circulating cell-free DNA inthe plasma of cancer patients during radiation therapyrdquo CancerScience vol 100 no 2 pp 303ndash309 2009
[44] A RThierry F Mouliere C Gongora et al ldquoOrigin and quan-tification of circulating DNA in mice with human colorectalcancer xenograftsrdquo Nucleic Acids Research vol 38 no 18 pp6159ndash6175 2010
[45] F Mouliere B Robert E Peyrotte et al ldquoHigh fragmentationcharacterizes tumour-derived circulating DNArdquo PLoS ONEvol 6 no 9 Article ID e23418 2011
[46] P Pinzani F Salvianti S Zaccara et al ldquoCirculating cell-free DNA in plasma of melanoma patients qualitative andquantitative considerationsrdquo Clinica Chimica Acta vol 412 no23-24 pp 2141ndash2145 2011
[47] H Lu J Busch M Jung et al ldquoDiagnostic and prognosticpotential of circulating cell-free genomic and mitochondrialDNA fragments in clear cell renal cell carcinoma patientsrdquoClinica Chimica Acta vol 452 pp 109ndash119 2016
[48] S Hauser T Zahalka J Ellinger et al ldquoCell-free circulatingDNA diagnostic value in patients with renal cell cancerrdquoAnticancer Research vol 30 no 7 pp 2785ndash2789 2010
[49] J Wan L Zhu Z Jiang and K Cheng ldquoMonitoring of plasmacell-free DNA in predicting postoperative recurrence of clearcell renal cell carcinomardquoUrologia Internationalis vol 91 no 3pp 273ndash278 2013
[50] M Fleischhacker and B Schmidt ldquoCirculating nucleic acids(CNAs) and cancermdasha surveyrdquo Biochimica et Biophysica Acta(BBA)mdashReviews on Cancer vol 1775 no 1 pp 181ndash232 2007
[51] V V Levenson ldquoDNA methylation as a universal biomarkerrdquoExpert Review of Molecular Diagnostics vol 10 no 4 pp 481ndash488 2010
[52] A M Dworkin T H-M Huang and A E Toland ldquoEpigeneticalterations in the breast implications for breast cancer detec-tion prognosis and treatmentrdquo Seminars in Cancer Biology vol19 no 3 pp 165ndash171 2009
[53] M O Hoque ldquoDNA methylation changes in prostate cancercurrent developments and future clinical implementationrdquoExpert Review of Molecular Diagnostics vol 9 no 3 pp 243ndash257 2009
[54] K Warton and G Samimi ldquoMethylation of cell-free circulatingDNA in the diagnosis of cancerrdquo Frontiers in Molecular Bio-sciences vol 2 article 13 2015
10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

10 Disease Markers
[55] P P Anglim T A Alonzo and I A Laird-Offringa ldquoDNAmethylation-based biomarkers for early detection of non-smallcell lung cancer an updaterdquoMolecular Cancer vol 7 article 812008
[56] Y Delpu P Cordelier W C Cho and J Torrisani ldquoDNAmethylation and cancer diagnosisrdquo International Journal ofMolecular Sciences vol 14 no 7 pp 15029ndash15058 2013
[57] J Charlton R D Williams MWeeks et al ldquoMethylome analy-sis identifies a Wilms tumor epigenetic biomarker detectable inbloodrdquo Genome Biology vol 15 no 8 article 434 2014
[58] T C Brown C C Juhlin J M Healy M L Prasad R Korahand T Carling ldquoFrequent silencing of RASSF1A via promotermethylation in follicular thyroid hyperplasia a potential earlyepigenetic susceptibility event in thyroid carcinogenesisrdquo JAMASurgery vol 149 no 11 pp 1146ndash1152 2014
[59] M K Joo K H Kim J-J Park et al ldquoCpG island pro-moter hypermethylation of Ras association domain family 1Agene contributes to gastric carcinogenesisrdquoMolecular MedicineReports vol 11 no 4 pp 3039ndash3046 2015
[60] X-M Wu Y Chen Y Shao X-L Zhou and W-R TangldquoAssociation between cigarette smoking and RASSF1A genepromoter hypermethylation in lung cancer patients a meta-analysisrdquo Asian Pacific Journal of Cancer Prevention vol 15 no19 pp 8451ndash8454 2014
[61] H-L Wang P Liu P-Y Zhou and Y Zhang ldquoPromotermethylation of the RASSF1A gene may contribute to colorectalcancer susceptibility a meta-analysis of cohort studiesrdquo Annalsof Human Genetics vol 78 no 3 pp 208ndash216 2014
[62] J-G Si Y-Y Su Y-H Han and R-H Chen ldquoRole of RASSF1Apromoter methylation in the pathogenesis of ovarian cancer ameta-analysisrdquo Genetic Testing and Molecular Biomarkers vol18 no 6 pp 394ndash402 2014
[63] K Daniunaite S Jarmalaite N Kalinauskaite et al ldquoPrognosticvalue of RASSF1 promoter methylation in prostate cancerrdquoTheJournal of Urology vol 192 no 6 pp 1849ndash1855 2014
[64] H Shi Y Li X Wang et al ldquoAssociation between RASSF1Apromoter methylation and ovarian cancer a meta-analysisrdquoPLoS ONE vol 8 no 10 Article ID e76787 2013
[65] M Spugnardi S Tommasi R Dammann G P Pfeifer and DS B Hoon ldquoEpigenetic inactivation of RAS association domainfamily protein 1 (RASSF1A) inmalignant cutaneousmelanomardquoCancer Research vol 63 no 7 pp 1639ndash1643 2003
[66] J Ellinger D Holl P Nuhn et al ldquoDNA hypermethylation inpapillary renal cell carcinomardquo BJU International vol 107 no4 pp 664ndash669 2011
[67] V V Gordiyuk G V Gerashchenko I Y Skrypkina etal ldquoIdentification of chromosome 3 epigenetic and geneticabnormalities and gene expression changes in ovarian cancerrdquoBiopolymers and Cell vol 24 no 4 pp 323ndash332 2008
[68] G V Gerashchenko V V Gordiyuk I Y Skrypkina etal ldquoScreening of epigenetic and genetic disturbances ofhuman chromosome 3 genes in colorectal cancerrdquo UkrainskiıBiokhimicheskiı Zhurnal vol 81 no 4 pp 81ndash87 2009
[69] M Kim J-H Kim H-R Jang et al ldquoLRRC3B encodinga leucine-rich repeat-containing protein is a putative tumorsuppressor gene in gastric cancerrdquo Cancer Research vol 68 no17 pp 7147ndash7155 2008
[70] A G Kondratov K A Nekrasov L V Lototska et al ldquoCom-parative analysis of epigenetic markers in plasma and tissue ofpatients with colorectal cancerrdquo Biopolymers and Cell vol 30no 2 pp 129ndash134 2014
[71] S Demokan A Y Chuang K M Pattani D Sidransky WKoch and J A Califano ldquoValidation of nucleolar protein 4 asa novel methylated tumor suppressor gene in head and neckcancerrdquo Oncology Reports vol 31 no 2 pp 1014ndash1020 2014
[72] Y Takada X Ye and S Simon ldquoThe integrinsrdquoGenome Biologyvol 8 no 5 article 215 2007
[73] A Mambole S Bigot D Baruch P Lesavre and L Halbwachs-Mecarelli ldquoHuman neutrophil integrin 12057291205731 up-regulation bycell activation and synergy with1205732 integrins during adhesion toendothelium under flowrdquo Journal of Leukocyte Biology vol 88no 2 pp 321ndash327 2010
[74] D G Stupack and D A Cheresh ldquoIntegrins and angiogenesisrdquoCurrent Topics in Developmental Biology vol 64 pp 207ndash2382004
[75] R Rathinam and S K Alahari ldquoImportant role of integrins inthe cancer biologyrdquo Cancer and Metastasis Reviews vol 29 no1 pp 223ndash237 2010
[76] A Ghosh S Ghosh G P Maiti et al ldquoFrequent alterationsof the candidate genes hMLH1 ITGA9 and RBSP3 in earlydysplastic lesions of head and neck clinical and prognosticsignificancerdquo Cancer Science vol 101 no 6 pp 1511ndash1520 2010
[77] L Hakkinen T Kainulainen T Salo R Grenman and HLarjava ldquoExpression of integrin 1205729 subunit and tenascin in oralleukoplakia lichen planus and squamous cell carcinomardquoOralDiseases vol 5 no 3 pp 210ndash217 1999
[78] S Roy L Bingle J FMarshall et al ldquoThe role of12057291205731 integrin inmodulating epithelial cell behaviourrdquo Journal of Oral Pathologyand Medicine vol 40 no 10 pp 755ndash761 2011
[79] A M Hoslashye J R Couchman U M Wewer K Fukami and AYoneda ldquoThe newcomer in the integrin family integrin 1205729 inbiology and cancerrdquo Advances in Biological Regulation vol 52no 2 pp 326ndash339 2012
[80] S Mitra D M Indra N Bhattacharya et al ldquoRBSP3 isfrequently altered in premalignant cervical lesions clinical andprognostic significancerdquo Genes Chromosomes and Cancer vol49 no 2 pp 155ndash170 2010
Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom

Submit your manuscripts athttpwwwhindawicom
Stem CellsInternational
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
MEDIATORSINFLAMMATION
of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Behavioural Neurology
EndocrinologyInternational Journal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Disease Markers
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
BioMed Research International
OncologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
PPAR Research
The Scientific World JournalHindawi Publishing Corporation httpwwwhindawicom Volume 2014
Immunology ResearchHindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Journal of
ObesityJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Computational and Mathematical Methods in Medicine
OphthalmologyJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Diabetes ResearchJournal of
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Research and TreatmentAIDS
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporationhttpwwwhindawicom Volume 2014
Parkinsonrsquos Disease
Evidence-Based Complementary and Alternative Medicine
Volume 2014Hindawi Publishing Corporationhttpwwwhindawicom