Localization of a gamma-glutamyl-transferase-related gene family on chromosome 22
-
Upload
christine-morris -
Category
Documents
-
view
213 -
download
1
Transcript of Localization of a gamma-glutamyl-transferase-related gene family on chromosome 22
Hum Genet (1993) 91:31-36 human .
genet,cs �9 Springer-Verlag 1993
Localization of a gamma-glutamyl-transferase-related gene family on chromosome 22
Christine Morris ~, C~line Courtay 2, 4, Ad Geurts van Kessel 3, Johanna ten Hoeve 4, Nora Heis terkamp 4, John Groffen 4
] Cytogenetic and Molecular Oncology Unit, Christchurch Hospital, Christchurch, New Zealand 2 Centre National de la Recherche Scientifique, Unit6 Associ6e 597, 30, Rue Lionnois, F-54000 Nancy, France 3 Department of Human Genetics, University Hospital Nijmegen, P.O. Box 9109, NL-6500 HB Nijmegen, The Netherlands 4 Section of Molecular Diagnosis, Department of Pathology, Children's Hospital Los Angeles, 4650 Sunset Boulevard, Los Angeles, CA 90027, USA
Received: 4 May 1992/Revised: 31 August 1992
Abstract. A gene family encompassing a minimum of four genes or pseudogenes for gamma-glutamyl trans- ferase (GGT; EC 2.3.2.2) is present on chromosome 22q11. We have previously isolated a cDNA related to GGT but clearly not belonging to its gene family. The chromosomal location of this related gene, GGTLA 1, has been determined by both isotopic and fluorescence in situ hybridization to metaphase cells and by Southern blot analysis of somatic cell hybrid DNAs. We show that GGTLA1 is part of a distinct gene family, which has at least four members (GGTLA1, GGTLA2, GGTLA3, GGTLA4). At least two loci are located on chromosome 22 within band q 11 and proximal to the chronic myeloge- nous leukemia (CML) breakpoint in BCR (breakpoint cluster region gene). At least one other member is located more distally between the breakpoints found in Ewings sarcoma and CML. Some of the GGT and GGTLA family members are located on N o t I restriction enzyme frag- ments of a similar size. Combined results indicate that a seg- ment of human chromosome 22q 11 has undergone large- scale amplification events relatively recently in evolution.
Introduction
Gamma-glu tamyl transferase (GGT; glutamine: D-glu- tamyl-peptide 5-glutamyltransferase; EC 2.3.2.2) is a key enzyme in the metabolism of glutathione. Most of the hu- man GGT cDNAs isolated to date appear to be transcripts from a single locus. However, a second mRNA transcript, which contains a different coding region and originates from a distinct locus, has been identified in kidney (Paw- lak et al. 1989). On a genomic level, multiple GGT genes or pseudogenes have been shown to exist. Pawlak et al. (1988) have molecularly cloned segments of four dif-
Correspondence to: J. Groffen
ferent loci. The amplification of GGT sequences has evi- dently occurred relatively recently in evolution (Heis- terkamp and Groffen 1988; Pawlak et al. 1988) as the rat genome contains a single-copy locus (Pawlak et al. 1988). In situ hybridization of a rat cDNA probe showed that GGT gene sequences were located proximally on human chromosome 22 at the interface of bands 22q11.1 and 22q11.2 (GGT1). A second signal at band q13.1 (GGT2) was also identified (Bulle et al. 1989). We have shown previously that one member of the GGT family is located distal to the BCR gene on chromosome 22ql 1, and lies ad- jacent to a BCR pseudogene locus (BCR-3). This GGT gene is actively transcribed and encodes the placental GGT cDNA (Heisterkamp and Groffen 1988; Rajpert-de Meyts et al. 1988). A second GGT-homologous region, again contiguous with a BCR pseudogene (BCR-4), was mapped centromeric of the functional BCR (BCR-1) gene (Heisterkamp and Groffen 1988). Recently we have re- ported the isolation of a cDNA encoding a protein similar to but distinct from GGT. The similarity between GGT and the newly identified gene product, is 39.5%. This pro- tein, initially designated GGT-rel, shares some important biological functions with GGT in that it is able to cleave the gamma-glutamyl moiety of some natural compounds that contain this group, such as leukotriene C4 and glu- tathione. However, it differs from GGT in that it is unable to cleave the gamma-glutamyl group of some synthetic substrates (Heisterkamp et al. 1991). Because of its GGT- like-activity, we now propose the name GGTLA1, which is accepted by Human Gene Mapping, for the locus en- coding this cDNA. Previous Southern blot analysis using small cDNA probes had suggested that GGTLA repre- sents a complex family of related genes (Heisterkamp et al. 1991). This was confirmed in the present study. In ad- dition, although the GGTLA complex is structurally dis- tinct from the GGT gene family, we show that most mem- bers of the two families are located within the same band, ql 1 on chromosome 22.
32
Materials and methods
Isolation o f GGTLA and G G T genomic clones
A small EcoRI-Sau3A fragment encompassing bases 1 188 of the 5" untranslated region of the GGTLA1 eDNA hpll4-1 (Heisterkamp et al. 1991) was used to isolate human genomic GGTLA clones from a partial MboI library constructed in a lambda bacteriophage vector from normal human leukocyte DNA (Clontech). The GGTLA 1.3-kb (kilobase) TaqI probe was isolated from genomic clone L8-1. This in- tron probe was devoid of repetitive sequences or plasmid sequences and did not hybridize to the GGTLAI cDNA. In addition, it showed no cross hybridization, under relaxed stringency washing conditions, to genomic GGT sequences isolated previously. Phage clone L7-1, used for fluorescence in situ hybridization studies, contained a 9.5-kb insert of genomic DNA and also showed homology only with GGTLA- specific probes. The GGT 0.45-kb BstEII probe was isolated from cosmid vKIL3, which contains the 3"end of the human GGT genomic locus (Heisterkamp and Groffen 1988). The probe originates immedi- ately 5" to the most 5" Sall site in VKll-3, it repeat free, and hy- bridizes with GGT loci.
DNA sources
DNA was isolated from a panel of somatic cell hybrid lines that con- tain part or all of chromosome 22. These included A3EW2-3B (Geurts van Kessel et al. 1985), X/22-33-11TG, and 1/22 AM 27 (Geurts van Kessel et al. 1980; Adham et al. 1989; Dumanski et al. 1990), WESP-2A-TG8, and WEGROTH D2 (Dumanski et al. 1990). Normal human and mouse DNA were prepared from peripheral blood leukocytes and a murine cell line, respectively. For field inversion gel electrophoresis (F1GE) studies, the cell line A172, derived from a glioblastoma, was obtained from the ATCC: lines TC71 and 5838 were obtained from Dr. T. Triche.
Gel electrophoresis, Southern blotting and hybridization
For standard Southern blots, somatic cell hybrid DNAs and human DNAs were subjected to electrophoresis on 0.7% agarose gels and blotted to nitrocellulose (Schleicher and Schuell, PH 79). For FIGE studies, DNA (6-10 lag) embedded in agarose blocks was digested overnight with 30 units of enzyme. Gels (1%, 250 ml, 20 • 25 cm) were preelectrophoresed for 30 rain at 200 V, and then for 24 h with a forward pulse increasing linearly from 3-120 s and an increasing backward pulse of 1-40 s. Gels were stained with ethidium bromide to localize markers (yeast chromosomes and lambda ladders), ex- posed to 302 mn UV for 10 rain to fragment high molecular weight DNA, then blotted to Nytran. Probes, isolated from vector, were la- beled by primer extension (Feinberg and Vogelstein 1984), and mem- branes were hybridized as described (Heisterkamp et al. 1983).
Isotopic and f luorescence in situ hybridization
Human metaphase cells were prepared from phytohemagglutinin (PHA)-stimulated peripheral blood of normal donors (male, 46,XY, and female, 46,XX), from the 46,XX lymphoblastoid cell line GM07387A (NIH Human Genetic Mutant Cell Repository) and from unstimulated leukemic cells of a patient with chronic myeloid leu- kemia, a breakpoint within M-Bcr of the BCR gene (Groffen et al. 1984; Heisterkamp et al. 1985) and a typical Ph' translocation, 46,XY, t(9;22)(q34;q22). Isotopic in situ hybridization of the 3H-labeled GGTLAI eDNA probe hp114-I to metaphase cells prepared from a karyotypically normal (46,XX) donor, and from the GM07387A cell line, was performed as previously described (Morris et al. 1990). Fluorescence in situ hybridization methods were essentially as de- scribed (Rowley et al. 1990) with minor modifications. The phage probe L7-1 was biotin-labeled by nick-translation using biotin-14- dATP (Bethesda Research Laboratories, Gaithersberg, Md). Alter RNase treatment, metaphase cells on slides were digested with pro- teinase K (0.2 lag/ml in 20 mM Tris-HC1, pH 7.5/2 mM CaC12) for 7.5 rain at 37~ rinsed three times, 2 min each, in 2 x SSC, pH 7.0, at room temperature, then fixed in 4% paraformaldehyde for 10 rain. Following a further three washes in 2 x SSC, chromosomal DNA was
denatured by immersion of the slides in 70% formamide/2 x SSC, pH 7.0 at 70~ for 2 min, and dehydrated in an ethanol gradient. Human Cotl DNA (Bethesda Research Laboratories, final concentration 250 ng/lal) was added to the hybridization mixture (50% fl~rmamide/2 • SSC/10% dextran sulfate/500 ng/lal sheared salmon sperm DNA), which contained approximately 2 ng/lal of labeled probe. After denab uration and pre-annealing steps, 20 lal of hybridization mix was ap- plied to each slide, the slides were incubated overnight ( 16 h) at 37~ washed, and the biotinylated probe was detected with fluorescein isothiocyanate (FITC)-conjugated avidin DCS (Vector Laboratories, Burlingame, Calif.), including signal amplification steps, as described (Rowley et al. 1990). Slides were mounted in glycerol containing the antifade agent p-phenylene diamine dihydrochloride (Sigma, I mg/ml), with 0.2 lag/ml propidium iodide as counterstain. Fluorescence mi- croscopy was performed with a Leitz Aristoplan microscope equipped with epifluorescence optics and a G/R filter block.
Results
Multiple GGTLA loci in the human genome
S o u t h e r n b lo t s tud ie s w e r e p e r f o r m e d to e s t a b l i s h the n u m b e r o f G G T L A loci p r e s e n t in the h u m a n g e n o m e .
c D N A p r o b e s w o u l d no t be s u i t a b l e fo r th i s t y p e o f a n a l y - sis b e c a u s e the i n t r o n - e x o n o r g a n i z a t i o n o f the h u m a n G G T L A I g e n e h a s no t b e e n d e t e r m i n e d . M u l t i p l e b a n d s m i g h t s i g n i fy the d e t e c t i o n o f d i f f e r en t e x o n s o f o n e g e n e ,
or r e p r e s e n t h o m o l o g o u s e x o n s o f d i f f e r e n t r e l a t ed genes . F o r th i s r e a s o n , g e n o m i c c l o n e s w e r e i so l a t ed f r o m a hu- m a n l i b r a ry u s i n g a v e r y sma l l ( 200 bp) 5" G G T L A 1 e D N A p r o b e ; a r e p e a t - f r e e 1 .3-kb TaqI g e n o m i c p r o b e
w as p r e p a r e d f r o m o n e o f t h e s e c l o n e s (see M a t e r i a l s a n d m e t h o d s ) . T h i s G G T L A in t ron p r o b e wil l no t h y b r i d i z e w i t h G G T loci . T h e 1 .3-kb TaqI p r o b e d e t e c t e d m u l t i p l e r e s t r i c t i o n e n z y m e f r a g m e n t s o f u n e q u a l h y b r i d i z a t i o n in- t en s i t y w i t h e v e r y e n z y m e t e s t ed in h u m a n g e n o m i c D N A (Fig. 1, l a n e s 1 -5 ) . F o r e x a m p l e , f ive d i s c r e t e EcoRI fYag-
m e n t s o f a p p r o x i m a t e l y 25, 20, 12, 10, a n d 2.5 kb hy -
Fig. 1. Southern blot analysis of gamma-glutamyl transferase like ac- tivity (GGTLA) copy number. DNA from an AML (acute myeloge- nous leukemia) patient was digested with EcoRI (lane l), HindIII (lane 2). Bglll (lane 3), BamHI (lane 4), and Sstl (lane 5). The posi- tion of lambda/HindllI molecular weight fragment markers is shown to the leJ?
33
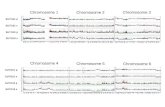
Fig.2 a-c. Chromosomal localization of the biotinylated GGTLA phage clone L7-1 after competitive in situ hybridization to metaphase cells and detection with FITC-conjugated avidin, a Control (46,XY) metaphase cell showing hybridization of L7-1 to two G-group chro- mosomes (arrows), identified as chromosome 22 homologues after G- banding (not shown); b partial early metaphase spread showing dif- fuse conglomerated signals on chromosome 22; c leukemic t(9;22) (q34;ql 1) metaphase cell showing hybridization of GGTLA L7-1 to chromosome 22 (right, lower arrow), close to the 9/22 breakpoint junction of the 9q+ derivative (right, upper arrow), and to the telom- eric end of the Ph chromosome long arm (left arrow)
bridized to the probe (Fig. 1, lane 1). Of these fragments, the 12-kb band hybridized most intensely. In an SstI di- gest, four bands of approximately 16, 12, 9, and 5.5 kb were visible, and the 12- and 5.5-kb bands were most in- tense (Fig. 1, lane 5). Strongly hybridizing bands may rep- resent two comigrating fragments with similar molecular weights. Because the probe does not contain an internal EcoRI or SstI site, each hybridizing fragment is likely to represent a discrete locus and we suggest that the GGTLA gene family encompasses approximately six members. Al- ternatively, hybridization intensity may reflect the degree of homology with the probe: the most closely related GGTLA species would hybridize more strongly than those with less homology and more distantly related. In this event, the GGTLA gene family would contain at least four members.
Chromosomal location of GGTLA
In situ hybridization using isotopic methods and the 3H- labeled GGTLA1 cDNA probe hpll4-1 showed strongest labeling proximally on chromosome 22q for both metaphase preparations tested. A combined total of 632 grains were scored on 200 metaphase cells: 94 grains (15%) were located on chromosome 22, and 77 of these (82%) were located in the region 22q l l -q12 , with a modal peak of 39 grains within band 22q11. Significant labeling was not detected on any other chromosome. This result was confirmed using FISH techniques and the
GGTLA-specific phage clone L7-1. In both normal hu- man male, 46,XY, and female, 46,XX, metaphase prepara- tions, an intense fluorescence signal was detected on the proximal q arm of two G-group chromosome in 8 0 % - 100% of the 20 cells analyzed (Fig. 2a, b). Subsequent destaining and G-banding of the same cells (after Canniz- zaro and Emanuel 1984) identified these as chromosome 22 homologues, and showed that the signal was confined within band 22q I 1 (not shown). In many early metaphases cells, where chromosomes are elongated, the signal ap- peared to represent a conglomerate of discrete hybridiza- tion sites (Fig. 2b). This observation, combined with the fact that significant labeling was not detected on any other chromosome, suggests that most GGTLA family members are located in very close proximity on chromosome 22. Hybridization of L7-1 to Ph-positive, t(9;22)(q34;ql 1) leukemic metaphase cells showed fluorescence signals on chromosome 22, on the telomeric end of the Ph chromo- some, and at the breakpoint junction of the 9q+ chromo- some (Fig. 2c). Chromosomes were identified by G-band- ing after hybridization analysis as described above. Spe- cific labeling on the Ph and on the 9q+ chromosomes is strong evidence for a minimum of two distinct loci. More- over, because the breakpoint on chromosome 22 occurred within M-Bcr, we can conclude that GGTLA family mem- bers are present telomeric and centromeric of the BCR gene.
Somatic cell hybrid analysis
To sublocalize these members more precisely and to con- firm our in situ hybridization data, Southern blot analysis on DNA from somatic cell hybrids containing different segments of chromosome 22 was performed. The restric- tion enzyme SstI was used for this purpose since digestion of human DNA yields four clearly identifable bands (16, 12, 9 and 5.5 kb) corresponding to the four (or more) loci. Some of the somatic cell hybrids used contain other chro- mosomes besides chromosome 22. However, hybrid WE- GROTH D2 contains chromosome 22 as its only human component (Dumanski et al. 1990). DNAs digested with
34
Fig.3. Localization of GGTLA using somatic cell hybrid DNAs. DNAs were digested with Sstl. Human DNA (lane 1); mouse DNA (lane 2); WEGROTH D2 (lane 3); A3EW2-3B (lane 4); 1/22 AM 27 (lane 5); X/22-33-IITG (lane 6); WESP-2A-TG8 (lane 7). Exposure time for lane 1 was 3h; for lanes 2-7, 48h. The approximate molecu- lar weights of the hybridizing fragments are as indicated to the right of the figure. Posthybridization washing was at a stringency of 0.01 x SSC, 65~
Ss t I were probed with the 1.3-kb Taql G G T L A genomic DNA fragment. Under the high-stringency post-hybridiza- tion washing conditions used in these experiments, mouse DNA showed no discrete hybridization signal (Fig. 3, lane 2). The hybrid W E G R O T H D2 contained the 16-, 12-, and 5.5-kb bands but lacked the 9-kb band (Fig. 3, lane 3; Fig. 5). Hybrid W E S P 2 A - T G 8 contains the 22q- chromosome generated by the Ph translocation in chronic myelogenous leukemia (CML). The 12- and 5.5-kb fragments were de- tected in WESP2A-TG8 (Fig. 3, lane 7; Fig. 5). Hybrid X / 2 2 - 3 3 - I I T G contains the q l l . l -q te r segment of chro- mosome 22 and also showed the presence of these bands (Fig. 3, lane 6; Fig. 5). This localized the 12- and 5.5-kb fragments distal to the X/22-33-11TG breakpoint in band q l l . 1 , but proximal to the CML breakpoint. The 16-kb fragment is absent from W E S P 2 A - T G 8 (Fig. 3, lane 4; Fig. 5) and is thus located distal to the C M L breakpoint. It is, however, proximal to the Ewings sarcoma breakpoint in band q12, because it was absent from somatic cell hy- brid A3EW2, which was derived from a Ewings sarcoma patient with a t(11;22) and contains the segment 2 2 q 1 2 - 22qter (see Fig. 3, lane 4). Consistent with this, the 16-, 12-, and 5.5-kb bands were present in hybrid 1/22AM 27, which contains the region 22pter-q13.1 (Fig. 3, lane 5; Fig. 5).
Fie ld invers ion gel e lec t rophores i s ( F I G E )
FIGE was used to further investigate and compare the structural organization of G G T L A and GGT family mem- bers. Hybridization with the GGT 0.45-kb B s t E I I intron genomic probe revealed a varying number o f N o t l frag- ments in different DNAs. For example, three fragments were seen in the cell line TC71 and five in the cell line 5838 (Fig. 4, lanes 5 and 6). The two smaller fragments o f 520 and 480 kb were consistently seen in all DNAs tested. Larger and smaller fragments in other DNAs may repre- sent differences in methylation, N o t l polymorphic sites, or both. When the same membrane was rehybridized with
Fig.4. F1GE analysis of GGT and GGTLA. DNAs were digested with Notl and hybridized to the 1.3-kb GGTLA genomic intron probe or the 0.45-kb GGT genomic intron probe. DNAs include those from the cell line A172, (lane I)," normal human leukocytes (hme 2); cell line TC71 (lanes 3 and 5); cell line 5838 (lanes 4 and 6). Lanes I 4 were hybridized to the GGTLA and lanes 5 6 to the GGT probe. The ap- proximate molecular weights of major common fragments are shown to the right of the panel. Note that lanes 1-2 are from a different gel than lanes 3 6 and that the DNA fragments have migrated less far
the G G T L A 1.3-kb Taql genomic probe, it was clear that many of the bands were common to both probes (Fig. 4, lanes 3 and 4). As with the GGT probe, the 520- and 480- kb bands were seen in all DNAs with GGTLA, but addi- tional fragments variable between samples were also pre- sent.
Discuss ion
GGT, a key enzyme in the metabolism of glutathione, was thought to be the only enzyme capable o f hydrolyzing the gamma-glutamyl bond of this compound. The isolation, characterization, and expression of the GGTLA1 cDNA showed that a second enzyme exists that has the same ca- pacity. It is possible that G G T L A was derived from one of the members of the GGT gene family by successive muta- tions in the course of evolution. However, we consider this possibility unlikely for two reasons. First, deduced amino acid sequences show considerable divergence between GGT and GGTLA. Secondly, the rat has a single-copy "genome equivalent" GGT gene and preliminary results suggest that the mouse genome also has only one G G T L A gene. Thus, GGT and G G T L A genes have existed inde- pendently for a relatively long time in evolution. In this context, the colocalization of GGT and G G T L A on chro- mosome 22 is somewhat surprising, as is the finding that both represent complex gene families.
One GGTLA-hybr id iz ing sequence represented by a 9- kb Ss t l fragment was absent from a chromosome 22-only somatic cell hybrid. This might suggest that the 9-kb band is not located on chromosome 22. We found it difficult to
35
3 5 7 4 6
I 1 6 1 6 - - 1 6
1 2 1 2 1 2 - 1 2
5.55.5,5.5 - 5.5
P
Q
12 - - -
1 1 . 2
11:; 1 1 . 2
1 2 . 1 1 2 . 2 1 2 3 ] a . ]
1 3 2
1 3 . 3
GGTLA* BCR-4
I IGLC I BCR-1 I'aoTLA ~BCR-3
22
Fig. 5. Schematic representation of the location of GGTLA gene fam- ily members. The approximate location of GGTLA gene family mem- bers with respect to IGlambda and BCR-1, -3 and -4 is shown to the right. The segments of chromosome 22 contained within the somatic cell hybrids is shown to the left. Numbers above the bars correspond to the lane numbers containing the different hybrid DNAs as indicated in Fig.3. The Sstl fragments found in each hybrid are listed beneath the bars. The asterisk denotes that two GGTLA loci (12- and 5.5-kb SstI fragments) are present in the region centromeric of the BCR-I gene
sociation between BCR, GGT, and G G T L A genes on 22ql 1 (Fig. 5) and suggests that these genes have been the site o f rearrangements resul t ing in the ampl i f ica t ion o f large seg- ments o f DNA, somet imes encompass ing comple te genes. One gene known to be involved in ch romosomal re- ar rangements is the BCR-1 gene, which is involved in t ranslocat ions in some forms o f human leukemia. How- ever, there is no evidence that this gene is genomica l ly un- stable. A second site of rearrangements is the l ambda im- munoglobu l in locus, which is also loca ted in this area (see Fig. 5). Al though this locus will rearrange in precursor B- cells to form a gene producing a funct ional l ambda chain, there is no evidence that the enzymes that media te these rear rangements are expressed in other cell types. And the past ampl i f ica t ion o f GGT, G G T L A , and BCR genes would have to have taken place in the germl ine in order to have been f ixed for posterity. Thus, the cause of this past large-scale ampl i f ica t ion event remains obscure. The ulti- mate result appears to have been the genera t ion o f a large number o f G G T and G G T L A genes. The indiv idual func- t ional status o f members of these complex gene famil ies remains to be determined.
reconci le this f inding with our ch romosome in situ hy- br id iza t ion results, which showed signif icant label ing on c h r o m o s o m e 22 only (F ig .2a , b). It is poss ib le that the probes used for in situ analysis were loca ted outs ide the area o f G G T L A h o m o l o g y detected by the 1.3-kb TaqI probe, or perhaps the region o f h o m o l o g y is too small to detect using in situ methodologies . It is also poss ib le that this f ragment contains an SstI po lymorph i sm. Al terna- t ively, a small region of ch romosome 22 has been dele ted in W E G R O T H D2. Fur ther work will be required to re- solve this issue.
The m i n i m u m of three other G G T L A loci, cor respond- ing to the SstI f ragments o f 16, 12 and 5 .5kb, could be m a p p e d using a combina t ion o f F I S H and somatic cell hy- brids. Two loci cor responding to the 12- and 5.5-kb SstI f ragments are loca ted cent romer ic to the C M L breakpoin t in band q l 1, whereas the other locus cor responding to the 16-kb f ragment is located in band q l l te lomer ic of the C M L breakpoin t (also see Fig. 5).
The G G T gene fami ly consists o f a m i n i m u m of four loci, all located on ch romosome 22. Of these, one fami ly m e m b e r is located adjacent to BCR-4 (see Fig .5) , a non- funct ional segment o f D N A with h o m o l o g y to the 3" end of the B C R gene (Heis te rkamp and Groffen 1988). BCR-4 is located cent romer ic to the funct ional BCR gene (BCR- 1), and is mos t p r o b a b l y e m b e d d e d be tween the im- m u n o g l o b u l i n l a m b d a cons tan t and var iab le reg ions (Croce et al. 1987; C. Morr is , unpubl i shed observat ions) . BCR-3, also nonfunct ional , is located te lomer ic o f BCR-1 and is phys ica l ly l inked to an ac t ively t ranscr ibed G G T gene fami ly m e m b e r (Heis te rkamp and Groffen 1988; see Fig. 5). Ch romosome 22 and q l 1 contains a m in imum of four G G T genes, three or more G G T L A genes, and four BCR genes. Some o f the G G T L A and G G T loci are lo- cated on NotI restr ic t ion enzyme f ragments o f ident ical size; the BCR-1 gene and one G G T gene are loca ted on the same NotI restr ict ion enzyme f ragment o f 4 8 0 k b (N. Heis te rkamp, unpubl i shed data). This indicates a c lose as-
Acknowledgements. We thank Jan-Willem Voncken for assistance; Lisa Uribe, Eva Knoppel, Miguel Brizuela, Margaret McDonald, and Suzanne Benjes for technical help; and K. C. Chen for photographic art work. This study was supported in part by National Cancer Insti- tute grant CA 47456 to J.G. and by grants from the Cancer Society of New Zealand and the Travis Trust to C.M.
References
Adham IM, Grzeschik K-H, Geurts van Kessel AHM, Engel W (1989) The gene encoding the human preproacrosin (ACR) maps to the ql3-qter region on chromosome 22. Hum Genet 88:59-62
Bulle F, Mattei MG, Siegrist S, Pawlak A, Passage E, Chobert MN, Laperche Y, Guella~n G (1987) Assignment of the human gamma- glutamyl transferase gene to the long ann of chromosome 22. Hum Genet 76 : 283-286
Cannizzaro EA, Emanuel B (1984) An improved method for G-band- ing chromosomes after in situ hybridization. Cytogenet Cell Genet 38:308-309
Croce CM, Huebner K, Isobe M, Fainstain E, Lifshitz B, Shtivelman E, Canaani E (1987) Mapping of four distinct BCR-related loci to chromosome region 22ql 1: order of BCR loci relative to chronic myelogenous leukemia and acute lymphoblastic leukemia break- points. Proc Natl Acad Sci USA 84:7174-7178
Dumanski JP, van Kessel AG, Ruttledge M, Sugawa N, Collins VP, Nordenskjold M (1990) Isolation of anonymous, polymorphic DNA fragments from human chromosome 22q12-qter. Hum Genet 84:219-222
Feinberg AP, Vogelstein B (1984) A technique for labeling DNA re- striction endonuclease fragments to high specific activity. Anal Biochem 13 : 266-267
Geurts van Kessel AHM, Westerveld A, De Groot PG, Meera Khan P, Hagemeijer A (1980) Regional localization of the genes coding for human ACO2, ARSA and NAGA on chromosome 22. Cytogenet Cell Genet 28:169-172
Geurts van Kessel AHM, Turc-Carel C, Klein A de, Grosveld G, Lenoir G, Bootsma D (1985) Translocation of oncogene c-sis from chromosome 22 to chromosome 11 in a Ewing sarcoma-derived cell line. Mol Cell Biol 5:427-429
Groffen J, Stephenson JR, Heisterkamp N, Klein A de, Bartram CR, Grosveld G (1984) Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell 36 : 93-99
Heisterkamp N, Groffen J (1988) Duplication of the BCR and gam- ma-glutamyl transpeptidase gene. Nucleic Acids Res 16:8045- 8056
36
Heisterkamp N, Groffen J, Stephenson JR (1983) The human v-abl homologue. J Mol Appl Genet 2 : 57 68
Heisterkamp N, Stam K, Groffen J, Klein A de, Grosveld G (1985) Structural organization of the BCR gene and its role in the Ph' translocation. Nature 315 : 758-761
Heisterkamp N, Knoppel E, Groffen J (1988) The first BCR gene in- tron contains breakpoints in Philadelphia chromosome positive leukemia. Nucleic Acids Res 16:10069-10081
Heisterkamp N, Rajpert de Meyts E, Uribe L, Forman HJ, Groffen J ( 1991 ) Identification of a novel human gamma-glutamyl cleaving enzyme related to gamma-glutamyl transpeptidase. Proc Natl Acad Sci USA 88 : 6303-6307
Morris C, Heisterkamp N, Hao Q L, Testa JR, Groffen J (1990) The human tyrosine kinase gene FER maps to chromosome 5 and is deleted in myeloid leukemias with a del(5q). Cytogenet Cell Genet 53 : 196-200
Pawlak A, Lahuna O, Bulle F, Suzuki A Ferry N, Siegrist S, Chikhi N, Chobert MN, Guella~n G, Laperche Y (1988) Gamma-glutamyl
transpeptidase: a single copy gene in the rat and a multigene family in the human. J Biol Chem 263 : 9913-9916
Pawlak A, Wu S-J, Bulle F, Suzuki A, Chikhi N, Ferry N, Balk J-H, Siegrist S, Guellaen G (1989) Different gamma-glutamyl transpep- tidase mRNAs are expressed in human liver and kidney. Biochem Biophys Res Commun 164 : 912-918
Rajpert de Meyts E, Heisterkamp N, Groffen J (1988) Cloning and nucleotide sequence of human gamma-glutamyl transpeptidase. Proc Natl Acad Sci USA 85 : 8840 8844
Rowley JD, Diaz MO, Espinosa R III, Patel YD, Van Melle E, Ziemin S, Tallon-Miller P, Lichter P, Evans GA, Kersey JH, Ward DC, Domer PH, Le Beau MM (1990) Mapping chromosome band 1 lq23 in human acute leukemia with biotinylated probes: identifi- cation of 11q23 translocation breakpoints with a yeast artificial chromosome. Proc Natl Acad Sci USA 87:9358-9362