DNA barcode sequence identification incorporating taxonomic hierarchy and within taxon variability
-
Upload
raunak-shrestha -
Category
Health & Medicine
-
view
445 -
download
4
Transcript of DNA barcode sequence identification incorporating taxonomic hierarchy and within taxon variability

Source:Little DP. DNA barcode sequence identification incorporating taxonomic hierarchy and within taxon variability. PLoS One. 2011;6(8):e20552.
Raunak Shrestha
13th Oct. 2011

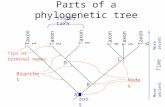
What is DNA Barcoding?
Barcoding is a standardized approach to identifying plants and animals by minimal sequences of DNA, called DNA barcodes.
DNA Barcode: A short DNA sequence, from a uniform locality on the genome, used for identifying species.
C A T G

DNA Barcoding developments
2003

DNA Barcoding developments (cont….)
2005
2007

DNA Barcoding developments (cont….)
2008
2009• MULTI-LOCUS GENE APPROACH FOR PLANT DNA BARCODING
• Chloroplast genes matK + rbcL recommended as the barcode regions
COI 1560 bp
BARCODE 648 bp
MINI-COI (186 bp)

Problems with conventional Sequence Identification Engines (SIDEs)
Source: Dr. F. Brinkman. Lecture slide-4 MBB741, 2011
SIDEs such as BLAST does not
consider Taxonomic
Hierarchy Information
Blastp Results

• Even a difference of single nucleotide can have significant impact on DNA Barcoding interpretation
• SIDEs such as BLAST and FASTA “corrects” it to overcome the sampling biasness.
• For closely related species, SIDEs such as BLAST and FASTA usually cannot diagnose such organism as separate species or of different taxon hierarchy
Problems with conventional Sequence Identification Engines (SIDEs) (cont….)
Character based Identification

Problems with conventional Sequence Identification Engines (SIDEs) (cont….)
• In a huge dataset using Parsimonous tree building method can generate large number of possible solution for even a small number of terminals
• “Computationally Expensive”
• Character-based phylogenetic methods requires multiple-sequence alignment (MSA).
• Several MSA tools may not be able to efficiently align the barcode sequences• Barcode sequence:
• Inter Species Variation > Intra-Species Variation • Conserved enough so that it could be amplified with ‘universal PCR
primers’ .
Phylogenetic Method based Identification

BRONX algorithm• BRONX (Barcode Recognition Obtained with Nucleotide
eXpose´s)• use an uncorrected character–based measure of similarity,• work with difficult to align markers, • capitalize upon knowledge of hierarchic evolutionary
relationships, • indicate ambiguous classification assignments, and• account for within taxon variation.

BRONX algorithm (cont…)• Reduces the reference sequences into a series of characters
defined by flanking context (‘pretext’ and ‘postext’)
The size of the pretext/postext used, and the range of text sizes stored, may vary by implementation.

BRONX algorithm (cont…)• Uses exhaustive tree construction algorithm• Then it starts comparing the sequences of each terminal
• Match the pretext and the postext of the paired sequences• If there is a pretext match as well as postext match
• Score for each combination shared with the paired sequences• If no match
• Determine all possible postext combination downstream of the matched pretext
• Choose the nearest postext match to the postext and align sequences accordingly
• Choose next postext and align the sequence• Score all the all alignment• The alignment with the highest final score is(are) considered
identification

Objective of the paper
To test the accuracy of BRONX sequence identification against leading published
SIDEs.

Dataset
• DNA Barcode sequence of matK and rbcL from databases• Sequences chosen only if both the sequences of matK and
rbcL were obtained from same individual (voucher specimen)• Global multiple sequence alignment• Alignment refined with MUSCLE• Sequence trimmed to be amplified with the following PCR
primers• matK 3F (5’-CGTACAGTACTTTTGTGTTTACGAG-3’) • matK 1R (5’-ACCCAGTCCATCTGGAAATCTTGGTTC-3’)
• rbcL aF (5’-ATGTCACCACAAACAGAGACTAAAGC-3)• rbcL aR (5’-GAAACGGTCTCTCCAACGCAT-3’)
• Final dataset: 2083 sequences of each marker representing 990 genera and 1745 species

Dataset
• Mini-barcodes:• Each of 2083 sequences were reduced to 100-200 base
sequences as the mini-barcodes.• Position of the barcodes were randomly chosen

Benchmarking• Benchmark of 11 different algorithms for both DNA barcodes
and mini-barcodes1. B = BRONX;2. C = CAOS; 3. D = DNA–BAR/degenbar;4. F = forced (constrained) tree–search; 5. J = SAP neighbor joining; 6. L = pairwise matching (local alignment); 7. N = NCBI-BLAST; 8. P = pairwise matching (global alignment); 9. S = SAP Barcoder; 10. T = de novo tree–search; 11. W = WU-BLAST.

Results
Genus-level identification
Weak test of species-level identification
Strong test of species-level identification
All test of species-level identification
Tests of identification using full–length barcode queries.

Results• Genus level identification highly successful (>99%) for BRONX,
DNA-BAR/degenbar, NCBI-BLAST and pairwise matching using full-length matK data
• rcbL not variable enough to distinguish between genera (~97% success)
• DNA-BAR/degenbar outperformed all other SIDEs in species-level identification • but BRONX too was significantly better in genus-level
identification
• BRONX should be preferred for genus-level identification queries over other SIDEs.

Results Tests of identification using mini-barcode queries.
Genus-level identification
Weak test of species-level identification
Strong test of species-level identification
All test of species-level identification

Results• For mini-barcode queries, identification success was relatively
lower than that of full-length queries
Identification success for strong test with combined matK and rbcL
Full-length query (DNA-BAR/degenbar)
Mini-barcode query (BRONX)
91 % 47 %
• Performance of DNA-BAR/degenbar was similar to other SIDEs for mini-barcode queries (11.24% success)
• Performance of BRONX for mini-barcode queries were better than all other SIDEs

• Moderate agreement among SIDEs for full-length queries (k=0.487-0.633)
• Little agreement among SIDEs for mini-barcode queries (k =0.191-0.137)
• Identification success did not improve with combined data of matk and rbcL.
Similarity of SIDE performance measured by Fleiss' index of interrater agreement (k)
Results

Conclusion• BRONX to be preferred over other SIDEs when
• Identification of genus are desired• Mini-barcode is used for identification
• DNA-BAR/degenbar exhibit superior performance in species level identification with full-length queries
• Due to inconstant performance no tree-based method should be used for barcode sequence identification
• BLAST is rapid means of sequence identification but other SIDEs provide better accuracy and consistency

Critique• Quality of sequence data in public database -> GIGO
• DNA barcode data depends upon the primer selected to amplify sequence• Use of only a single primer set of each locus• Does this mimic the real world dataset ?
• It would have been even better if the performance was measured in terms of computing time required for analysis.
• It seems that, till date, no algorithm is available which can incorporate both full-length query sequence as well as mini-barcode sequence query and give higher identification success at both genus and species level identification.

Questions ?
Thank you









![Untitled-1 [] · taxonomic characters. Zoological Nomenclature, origin of code, ICZN. UNIT V : Taxonomic Records And Publications Taxonomic keys, Taxonomic characters description,](https://static.fdocuments.us/doc/165x107/5e60819b810ee55ab507dd7d/untitled-1-taxonomic-characters-zoological-nomenclature-origin-of-code-iczn.jpg)