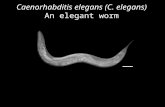
C. elegans was the first animal to have its genome (10 8 bp) completely sequenced.
-
Upload
hope-woodhams -
Category
Documents
-
view
218 -
download
0
Transcript of C. elegans was the first animal to have its genome (10 8 bp) completely sequenced.
C. elegans Genome in Numbers
Size(Mb)
Proteingenes
Protein genes/
kb
CodingRegions (%)
% of genes EST
matches(Mb)
95.53 19,141
4.98 27 38.9
The completed genome sequence is made up of
2,527 cosmids
113 fosmids
257 YACs
44 long range PCR products
intergenic DNA 47%
exonic DNA 27%
intronic DNA 26%
http://nema.cap.ed.ac.uk/Caenorhabditis/C_elegans.html
Characteristics of the Caenorhabditis elegans genomeSeq
GenesEST’sLev
Inv. Rep.
Tand RepTelom
The C. elegans genome consortium, TIG 15:51, 1999
The study of many aspects of Nematode biology were made easier after the genome of C. elegans was sequenced
After doing “discovery science” (L. Hood, 2002; “defining the elements in biological objects”) we can do regular “hypothesis-driven” science.
Banerjee & Slack BioEssays, 24:119, 2002
Involvement of small RNAs in the control of Temporal development and RNAi in C. elegans
24h
48h
72h
96h
Bacteria with no plasmid
Kamath e cols. (2001) Genome Biol. 2:1-10
Feeding C. elegans with dsRNA turns off specific genes
Other nematodes help define what is common to all species and what is restricted to C. elegans at the molecular level
Made by R. Turner Department of Biology, 1990 - Indiana University
CEW1 C. elegans
Anterior
Left side
SEM of CEW1 and C. elegans
Although the final morphology is the same, the induction events during vulva development are
difrerent in C. elegans and CEW1
Dichtel et al., Genetics, 157:183, 2001
Introns of the C. elegans vitellogenin genes
Winter, in: “Reproductive Biology of Invertebrates,Volume XII” - 2002
NJ-Tree of nematode vitellogenins (aminoacid sequences)
Winter, in: “Reproductive Biology of Invertebrates,Volume XII” - 2002
N- and C-terminus of nematode VTG evolve at different rates
Winter et al., Mol. Biol. Evol., 13:674, 1996
CUU
CUC
CUA
CUG
51 24
39 55
1 2
0 12CCA
CCC 1
99
64
985 17
15 83
CAA
CAG
GAU
GAC
50
50
23
77
Ce CEW1 CEW1Ce Ce CEW1 Ce CEW1
Codon usage table of vit-6 genes in C. elegans and CEW1
Winter et al., Mol. Biol. Evol., 13:674, 1996
Acknowledgements
Dr Cristiane Penha-Scarabotto
Dr Rubens Nobumoto AkamineJoselene Pereira de MouraIl-Young Ahn
Juliana Machado AndreoniPaulo Afonso de Carvalho
Daniela Peres Almenara
Manoel Aparecido Peres
SUPPORTED BY &
Undergraduatestudents
“... We found that bringing together the true cross-disciplinary scientists was rendered difficult by the fact that our academic center, and most academic centers, live in a world of departments. And the departments tend to create barriers both in how their students are educated and what the expectations are for faculty...” Leroy Hood, JAN/18/2002
[see http://www.oreillynet.com/pub/a/network/2002/01/18/hood.html]
http://www.oreillynet.com/biocon2002/?xA;
Wormbase
How are the genome sequences of C. elegans stored
Published in: http://nema.cap.ed.ac.uk/Caenorhabditis/C_elegans_genome/Celegansinformatics.html
Distribution of repetitive elements on the C. elegans chromosomes
Surzycki and William R. Belknap, PNAS 97:245–249, 2000
Cel
e 14
Cel
e 2
Cel
e 1
MIT
E-li
keC
ele
42
Saigusa et al., Curr. Biol. 12(2):R46, 2002
Movement
Kippert et al., Curr. Biol. 12(2):R47, 2002
Osmotic Shock Resistance
C. elegans behaviour is subjected to circadian variations
Caenorhabditis elegans vitellogenin genes
gene intron nr.
location
DNA stran
d
"YP"
Size of the
vitellogenin (kDa)
(*)
cDNA clones
(**)
chromosome: position
vit-1 4 K09F5.2
+ 170B
186.6 17 X: 7,742,201-7,746,243
vit-2 4 C42D8.2
+ 170B
186.2 118 X: 5,114,376-5,119,411
vit-3 5 F59D8.1
- 170A
185.0 1 X: 3,626,224-3,612,142
vit-4 5 F59D8.2
- 170A
184.8 5 X: 3,618,023-3,612,577
vit-5 5 C04F6.1
+ 170A
184.9 69 X: 3,416,833-3,421,907
vit-6 4 K07H8.6
+ 115/88
191.0 126 IV: 8,273,968-8,279,162
* = calculated from the amino acid sequence deduced from the gene sequence; signal peptide excluded.** = Number of cDNA clones listed in WormBase (http://www.wormbase.org/) (Jan/30/2001) associated with each gene
Winter, in: “Reproductive Biology of Invertebrates, Volume XII” - 2002
Embriogenesis of Acrobeloides nanus
Schierenberg (1999) - http://www.uni-koeln.de/math-nat-fak/zoologie/agschier/abschier3e.html