Basic Molecular Approaches: Cloning and Beyond...1 Basic Molecular Approaches: Cloning and Beyond...
Transcript of Basic Molecular Approaches: Cloning and Beyond...1 Basic Molecular Approaches: Cloning and Beyond...

1
Basic Molecular Approaches: Cloning and Beyond
PHOP 6241/6242 Fall 2018 Deborah Otteson, Ph.D.
UH College of Optometry
2

2
The Human Genome Project: What It Means for You
Genetics is becoming an important factor in the diagnosis and treatment of all kinds of disorders.
4
Applications of Molecular Biology and Genetics in Medicine
• Understand processes regulate normal and abnormal function incells and tissues
• Identify and understand genetic factors that cause disease
• Predict susceptible individuals/prevention
• Develop gene replacement/corrective therapies
• Identify new drug/treatment targets
• Identify patients who will benefit from different therapies (differentcauses = different treatments)

3
5
Developing New Treatments: Understand the Biological Processes Involved in Disease
ARMDGenetic/environmental Death of photoreceptors Defects in RPE Excessive vascular growth
Dry Eye
Genetic/environmental
Nerve damage
Defects in tear production
Hormonal
KeratoconusGenetic/environmental Disintegration of Bowman’s membrane Excessive protease activity
Glaucoma
Genetic components
Death of ganglion cells
Increased IOP
Defects in aqueous production/outflow
Comorbidity (diabetes?)
6
What is Molecular Biology?
A powerful approach for studying
• Cellular/biological processes at the molecular level• Biochemical processes within biological systems
• Molecular Biology tends to be “gene” based

4
“Central Dogma” of Gene Expression and Protein Synthesis
2-7
RNA viruses: Reverse transcription(e.g. HIV, retroviruses)
2-8
Chromosomes
• Visible at the light microscopic level in cells
during mitosis
• First named in 1888
• stained by ‘new’ synthetic chemical dyes
• chroma, "color" and soma, "body”
• Number of chromosomes varies between
species.
Human: Diploid = 46 chromosomes
(23 from each parent)22 autosomes
X & Y (sex chromosomes)
Examples of Diploid chromosome #:
Flies: 8 Mice: 40
Frogs: 26 Duck: 80
Asparagus: 30 Armadillo: 64
Yeast: 32 Sweet Potato: 90
Fluorescently stainedHuman chromosomes

5
2-9
Chromosomes contain Genes
• The term ‘gene’ was coined in 1909 as the
“unit of genetic inheritance”
• VERY Controversial at the time
• 1933: “There is no consensus opinion
amongst geneticists as to what the
genes are—whether they are real or
purely fictitious.” T. H. Morgan
• 2017: We now know that the genes are
genetic units in the DNA that code for
the RNAs and proteins necessary for
cell function and structure
• Humans: 20-25K protein coding genes
in genome
Arrows: Fluorescently labeled genes in condensed human chromosomes
2-10
Chemical Composition of Nucleic Acids
DNA consists of 4 nitrogen-containing bases
Purines first synthesized in laboratory: ~1888
base nucleoside nucleotideadenine adenosine dATPguanine guanosine dGTP
Pyrimidines first synthesized in laboratory: ~1879
base nucleoside nucleotidecytosine cytidine dCTPthymine thymidine dTTP
Base + deoxyribose = nucleosideBase + phospho-deoxyribose = nucleotide (= nucleoside with phosphate)

6
Nucleotides = Deoxyribose sugar + Base (Purine or Pyrimidine) + Phosphate group
2-11
Deoxyribose
5
4 1
3 2
Carbon atoms of ribose molecules are numbered
Base attaches at #1 carbon
Deoxyribose lacks oxygen molecule on #2 carbon (H instead of OH)
Phosphate groups attached to #5 carbon
5’ and 3’ carbons are where nucleotides are joined during DNA synthesis (more on this later)

7
Watson and Crick deduced the structure of DNA using data from Franklin and Wilkins (1953)
2-13
X-ray crystallography data
• Helical DNA structure
• Base pairing: fit previously
• Determined ‘rules’ of DNA
composition (A=T, C=G)
X‐ray diffraction photograph of DNA fiber
at high humidity (Franklin and Gosling,
1953b). Interpretation of the helical‐X and
layer lines added in blue.
Watson‐Crick model of DNA, adopted from
(Watson and Crick, 1953b), with the helical
repeat associated with the layer lines
labeled. Image from: DNA Structure:
Alphabet Soup for the Cellular Soul
2-14
Watson, Crick and Wilkins:Nobel Prize in Physiology for Medicine 1962
Watson and Crick with model of double helix
(very short paper published in Nature, 1953)

8
2-15
Double-stranded alpha helix
One full turn of helix ~10 base pairs
DNA base pairs more exposed at major groove
Regulatory proteins preferentially bind to major groove (more on thislater)
majorgroove
minorgroove
The structure of DNA
3.4Å
34Å
Watson-Crick Pairing
Invariate pairing bases
• Each base pair consists of
• 1 purine and 1 pyrimidine
T – A2 hydrogen bonds
C – G3 hydrogen bonds
Pyrimidines Purines
Hydrogen bonds

9
DNA: Primary Structure
Anti‐parallel orientation of 2 strands
5’‐CAGTGATTACA‐3’3’‐GTCACTAATGT‐5’
Phosphodiester bonds between deoxyribose sugars on backbone
= nucleotide
2-18
Semiconservative DNA replication
Meselson and Stahl, 1958
• Each parent strand serves as thetemplate for synthesis of a new daughter strand
• Invariant base pairing keeps sequencethe same in new strand
• Newly synthesized strand is basepaired with a parent strand
• During mitosis, each daughter cellreceives chromosomes each containing one original and one new strand of DNA

10
2-19
How much DNA in a human?
Human cells contain23 pairs of chromosomes
20,000-25,000 genes
6 x 109 base pairs
~ 2 meters of DNA in one human CELL(if it could be stretched out !)
Total length of DNA in ONE human(> 10 trillion cells)
~20 trillion meters~ 20 billion km
Distance: earth to sun: ~150 million km
One person’s DNA would go from earth to sun and back ~66.7 times!!!!
These calculations don’t include DNA from:
Mitochondria Microbes in/on the body
2-20
Forms a bead-like structure (11 nanometer diameter) = chromatin
Chromatin condenses into 30 nm fibers
Eukaryotic DNA: Higher Order Structure
DNA double helix coils around the histonehetero-octomer (2 times, 146 bps) = nucleosome
Histone modifications (acetylation and methylation) regulate chromatin packing

11
2-21
Chromatin is organized onto protein scaffold
Packing density increases in mitotic chromosomes
Smallest human chromosome is 2 micrometers long when condensed.
Eukaryotic DNA Higher Order Structure
3-22
Transcription: DNA to RNA
• Transcription occurs in both the nucleus and mitochondria• Genomic organization and transcription/translation processes in
mitochondria are similar to prokaryotic cellsFigure 1-13

12
3-23
RNA vs DNA
RNA DNASingle stranded Double stranded
Anti-parallel strands
A, C, G, U A, C, G, TRibose sugar Deoxyribose sugar
5’ 3’ 5’ 3’
5’3’
Sense strand/non-template
Anti-sense strand/template
(RNA) (DNA)
3-24

13
3-25
Classes of RNA
Protein coding RNAs: Messenger RNA (mRNA)
• transcribed from genes in DNA
• used as template for protein synthesis
Functional RNAs : tRNA, rRNA, snRNA, miRNA
• not translated into protein
• RNA is active component
Genomic RNA
• Retroviral genome
3-26
Functional RNAs
Transfer RNA (tRNA)
• brings the amino acid to the ribosome during proteinsynthesis (translation)
Ribosomal RNAs (rRNAs)
• components of the ribosomes
Small nuclear RNAs (snRNAs)
• multiple functions
• guide the modification of mRNA
• combine with proteins to form the ribonucleoproteinprocessing complex (spliceosome)

14
3-27
RNA regulation of gene expression:
Micro RNA (miRNA)
• Encoded in genome (can be individual miRNA genesor within introns of protein coding genes)
• Contains complementary sequences to mRNA target
• Bind to the 5’ untranslated region of mRNA target
• Targets RNA for degradation by cell
Small interfering RNA (RNAi)
• Anti-viral mechanism in lower eukaryotes
• RNA binds to the mRNA at the translation start site
• Blocks initiation of translation/protein synthesis
• Used in research to alter gene expression
More Functional RNAs
RNA transcription and protein synthesis take place in different parts of the cell
Griffiths. Figure 8-11
• RNA transcription• The generation of RNA from a DNAtemplate• Occurs in the nucleus•Regulated process
transcription factorsRNA polymerasechromatin structure
Protein coding RNAs are processed in nucleus:
• capping• splicing• poly‐adenylation
Processed mRNAs are transported out of the nucleus
Protein synthesis (translation) occurs in cytoplasm

15
Adjacent genes can have different orientations in the chromosome
3-29
Coding strand for gene 1
Coding strand for gene 3
Template strand for gene3
3-30
Eukaryotes: Gene Organization
Promoter region:• binding sites in DNA sequence interact with:
• RNA polymerase• transcription factors• Other DNA modifying enzymes
• +1 is transcription start site (TSS)• Bases in DNA sequence are numbered relative to TSS
• promoter starts at -1 and goes to left (-1, -2, -3 etc)• transcribed region starts at +1 and goes to right

16
3-31
Eukaryotes: Gene Organization
Introns = transcribed but removed during splicingExons = transcribed; remain in mRNA after splicing
Exons include both untranslated and protein coding regions
Note: Prokaryotic genes do not have introns
DNA condensedheterochromatin
Densely packed nucleosomes
Histone modifications stabilize structure (deacetylation, methylation)
DNA methylation at CpG sequences can permanently shut down expression
Transcriptional regulatory are proteins excluded from heterochromatin
DNA open = euchromatin

17
DNA condensed =heterochromatin
DNA open = euchromatin
Histones moved aside (sliding and/or disassembly)
Histone modifications open the DNA structure (acetylation)
Allows transcriptional regulatory proteins to bind to DNA
Transcription factors can recruit histone modifying enzymes RNA POLYMERASE and other components of the transcription machinery
5-34
Histones Influence Transcription
Histones: Eukaryotes
octamer containing two copies of 4 subunits
DNA wraps histone ~1.65 times to form nucleosome
(~147 base pairs)
Histone tails exposed
Modification of histone tails regulates chromatin structure (open vs. closed)

18
• RNA polymerase is recruited to the basal promoter by various
transcription factors bound to the proximal and distal promoter
• RNA Polymerase transcribes both Introns and Exons
• Results in hnRNA = heterogeneous nuclear RNA = (pre-mRNA)
Eukaryotes: Transcription Overview
hnRNA
3-36
RNA is Synthesized 5’ to 3’ Direction
• The first ribonucleotide of new RNA is the 5’ end of the RNA
molecule
• RNA is the reverse complement of template (anti-sense)
strand
• RNA sequence is same as non-template (sense) strand
• EXCEPT: RNA has U instead of T
Direction of RNA synthesis

19
3-37
RNA is Synthesized 5’ to 3’ Direction
• Template DNA strand copied from 3’ to 5’
• RNA synthesized from 5’ to 3’
• New bases are added to the 3’ end of the growing RNA
• Note that two DNA strands rejoin into double helix
3-38
Eukaryotes: RNA Processing in nucleus
Capping protects 5’ end
Splicing removes introns
AAAAAAddition of poly A tail stablilizes RNA
and signals for nuclear export

20
RNA Processing: Alternative Splicing
Multiple protein isoforms generated from a single geneDifferential exon usageRegulation of timing and tissue specificity of gene expressionCan have different functions
3-40
Eukaryotes: RNA Processing in nucleus
AAAAA
3-40
CODING REGIONUntranslated regions = 5’ UTR 3’UTR
Mature mRNA
Export from nucleus
Translation into proteins

21
4-41
Translation: RNA to Protein
4-42
20 Amino acids: Classified by structure and chemical composition
Non-polar, aliphathic (non polar, hydrophobic)Alanine Ala Glycine Gly Polar, unchargedIsoleucine Ile Asparagine Asp Leucine Leu Cysteine CysMethionine Met Glutamine GlnValine Val Proline Pro
Serine SerNegative charge Threonine ThrAspartate Asp(Aspartic Acid) Aromatic (C- ring)Glutamate Glu Phenalanine Phe(Glutamic Acid) Tryptophan Trp
Tyrosine TyrPositive chargeArginine Arg Histidine His Lysine Lys

22
4-43
• Each amino acid called “residue” (R)
• Each AA is numbered: starting at #1 at N terminal (amino end), withsequential numbering to C-terminal end.
• Co-linearity of DNA, RNA and protein sequence
• Amino acids linked by peptide bonds
• Multiple amino acids linked together form a polypeptide
• Properties of amino acid side chains confer structure and function ofprotein
Levels of protein structure: Primary
Polypeptides can bend into
regularly repeating structures
created by H-bonds between
-COOH and -NH2 groups of
different amino acid residues
4-44
Levels of protein structure: Secondary

23
4-45
Tertiary structure = 3-dimensional architecture
• Chaperon proteins in ER can regulateprotein folding.
• Proteins that are incorrectly folded arerefolded or degraded
• Stabilized by various intermolecularbonds:
• disulfide
• electrostatic
• hydrogen
• van der Waals
• Amino acids located far apart inprimary sequence can be brought close together by folding
Myoglobin
Levels of protein structure: Tertiary
4-46
Many mature proteins are composed of multiple subunits = Multimeric
Joined by various intramolecular bonds
Subunits: separate proteins, may be encoded by the same or different genes
# subunitsDimer 2Trimer 3Tetramer 4
Homodimer: 2 identical subunitsHeterodimer: 2 different subunits
Example of a tetramerHemoglobin
= 2 each of 2 subunits
Levels of protein structure: Quaternary

24
4-47
How is mRNA Sequence Translated into Amino Acids?
Genetic Code:
•Amino acids are encoded
by base triplets called
codons
• Many amino acids are
encoded by more than one
codon (degenerate)
• Codons specifying the
same amino acids can
differ at the 3rd base, known
as wobble position
Translation: RNA to Protein
Genetic CodeAmino acids are encoded by base triplets called codons
Many amino acids are encoded by more than one codon (degenerate)
Codons specifying the same amino acids differ at the 3rd
base known as wobble position

25
4-49
Compare Overlapping vs. Non-Overlapping Code
If overlapping, change (mutation) of one base could alter 3 codons
If non-overlapping, only one codon affected
Which one is actually used? Non-overlapping
Consequences of point mutations
Silent mutations: no change in protein
Missense: consequence depends on location and similarity of new amino acid to original
Alters multiple amino acids: consequences varypremature termination; loss of function, gain of function
Can alter gene expression if in regulatory regionMay be silent if not in critical region

26
4-51
Components for Translation
mRNA messenger RNA
Amino Acids
tRNAs = transfer RNA
Ribosomes
rRNA = ribosomal RNA
ribosomal proteins
tRNA
Multiple tRNAs with different
structures and anticodons
Enzymes couple amino acid to
correct tRNA molecule
Anticodon arm base‐pairs with
mRNA
Provides sequence specificity
for translation

27
Two Subunits of a Ribosome Contain
Protein and RNA Molecules
• Two ribosomal subunits
• Both contain rRNA and proteins
• Small subunit binds mRNA cap
• Scans mRNA to find translationstart site:
• identified by Kozak sequencein mRNA and AUG(methionine) start codon
• Recruits large subunit
• Translation begins
4-54
1. Initiation complex assembles at 5’ capof mRNA• 40S subunity• Met-tRNA• Initiation factors +GTP
2. Initiation complex scans 5’ end of mRNAfor KOZAK sequence and start codon
(Marilyn Kozak, 1987)
-6 +15’- GCCA/GCCAUGG-3’
3. Positions Met tRNA at AUG
Translation in Eukaryotes: Initiation
Figure 9-15

28
Figure 9-13
Key Sites of Interaction in the Ribosome
A=Amino acyl site: binds aminoacyl tRNA
P=Peptidyl site: forms peptide bond
between peptide chain and new amino acid
E= Exit site: tRNA without amino acid is released
Path of each tRNA through ribosome:A P E
4-56
Termination similar in pro- and eukaryotes
Ribosome reaches a stop codon (at A site)
Release factor (RF1) binds
There is no tRNA for STOP codon: only release factor
Disassembly of thetRNA-ribosome-mRNA complex
Peptidyl cleavage to release of nascent polypeptide from tRNA
Translation: Termination

29
4-57
Post-Translational Processing of Proteins
Protein processing begins in trans-golgi network
FoldingChaperone proteins regulate folding (see tertiary structure)
Proteolytic CleavageProteins synthesized as inactive precursorCleaved to generate active forme.g. pre-proinsulin > proinsulin > insulin
Glycosylation
Covalent attachment of carbohydrates (sugars)O-linked attached to AA with hydroxyl groupsN-linked attached to AA with amine groupsOften associated with recognition/adhesive function
4-58
Post-Translational Processing of Proteins
Methylation addition of a methyl group (CH3) e.g. calmodulin, Cytochrome C
Acetylation C-terminal acetyl group, allows addition of myristol group for membrane anchoring;
e.g. catalytic subunit of cyclic AMP-dependent protein kinase (PKA) is myristoylated
Phosphorylation often associated with intracellular signal transduction
e.g. Ligand binding to a receptor causes phosphorylation (auto-phosphorylation of receptor and/or phosphorylation of G-protein) and initiates intracellular signaling)

30
Protein Targeting: Signal sequences target proteins for secretion
4-59
Signal sequence: series of hydrophobic amino acids at N terminus of polypeptide
Targets assembled ribosome to the rough endoplasmic reticulum
Signal sequence is cleaved for secreted proteins
Most transmembrane proteins targeted to rough ER by hydrophobic transmembrane domain—not cleaved
4-60
Protein Targeting/Subcellular Localization
Nuclear Localization Signal directs protein to the nucleus. e.g. transcription factors, histones
Signal Sequence sends ribosome with new/growing polypeptide to endoplasmic reticulum. Protein is synthesized into the endoplasmic reticulum.
secreted proteins e.g. insulin, collagen.
Transmembrane proteins have signal sequences but not cleaved
other proteins destined for vesicles have signal sequences
Ubiquitination attaches Ubiquitin peptide; identified proteins for destruction; sends proteins to proteosome/endosomes

31
4-61
Protein Targeting: Ubiquitinization Targets a Protein for Degradation

32
Recomb DNA 63
Isolation of specific DNA or RNA/cDNA
• Diagnosis
• Nucleotide sequencing/gene ‘chip’ analysis; GWAS
• Identify and understand effects of mutations
•Probes for analysis of mutation or gene expression
• Transfer genes into cells/organisms for functional analysis or gene
therapy
• Generate recombinant proteins for research or therapeutics
(e.g. recombinant insulin, recombinant growth hormone)
Uses of Molecular Cloning
2-64
Transcriptome:The collection of all RNA transcripts in a cell.
The genome is identical in all
cells
Proteome: Collection of all proteins in a cell
or tissueGriffiths 10th editionFigs. 1-2 & 1-5

33
Identify the Gene Mutated in Patients with Eye Disease
Example: Age Related Macular Degeneration
Genetic studies can localize chromosomal regions associated with phenotypeNow need to identify candidate genes
‐genes that are expressed in affected tissue (retina, choroid and vasculature)
‐genes that are involved in functions affected tissue (eye development, cellular differentiation, phototransduction, angiogenesis, inflammation ???)
Online Genome DatabasesNational Library of Medicine/NIH
http://www.ncbi.nlm.nih.gov/genome/guide/human/
University of California Santa Cruz http://genome.ucsc.edu/
Ensembl http://www.ensembl.org
• GWAS or other genetic methodslocalize region associated withdisease
• Identify genes that would be likelycandidate for causing disease
• Sequence the DNA to look forpossible mutations
• Use Bioinformatics analysis of humangenome databases
• Analyze mRNA and proteinexpression
• Determine protein function
• Determine effect of mutations: whydoes it result in disease?

34
Isolation of Genomic DNA for Cloning and Analysis
Needed for genetic analysis, identification of mutation and variations
Sources of DNA
• Isolate DNA frompatient/subject– Blood
– Cheek swab
– Biopsy (e.g. cancer)
• DNA in all cells shouldbe the same **
** But see article in New York Times 9/24/13: Mosaicism in human genomeshttp://www.nytimes.com/2013/09/17/science/dna-double-take.html?pagewanted=all&_r=0
http://www.alleight.com/Products/Epicentre/BuccalAmp.htm
Agarose Gel Electrophoresis of DNA/RNA
http://www.stanford.edu/group/hopes/diagnsis/gentest/f_s02gelelect.gif
pole

35
Recomb DNA 69
Cloning and Analysis of RNA
Needed for:• identification of genes expressed in a
particular tissue/cell type.
• effects of mutation on size and stability ofmRNA
• analysis of consequences of mutation onproteins/cellular function
• cloning genes for gene replacementtherapy
Ethidium Bromide stainedAgarose gel of total RNA
28S and 18S RibosomalRNA
5S Ribosomalt-RNAmicroRNAs
mRNA
RNA Sources:• Tissue of interest
• Synthetic RNA (by machine)
• In vitro transcribed (from cloned DNA)
Recomb DNA 70
Genes and Gene Products for Analysis
Capping, Splicing Polyadenylation
CODING REGION
Protein
mRNA
cDNA: complementary DNA(copy of mRNA)
Genomic DNA
AAAA
AAAA

36
Recomb DNA 71
Analysis of RNA:Reverse Transcription Generates cDNA Copy
• Isolate total RNA from cells
• Oligo-dT primers bind to poly-A tailof mRNA
• Reverse transcriptase enzymemakes DNA copy of RNA
• RNAse removes RNA strand
• DNA polymerase synthesizessecond strand
• Results in double-stranded cDNA(complementary DNA); includescopies of all mRNAs present inoriginal sample
Analysis of Gene Expression: RT-PCR
RT = reverse transcription
PCR = polymerase chain reaction
RNA cDNA PCR with gene specific primers gel electrophoresis
Standard RT‐PCR
end point assay: gene is present/absent
qualitative, not quantitative
Quantitative RT‐PCR : “real time”
determine copy number or relative abundance of RNA (cDNA)
(e.g. compare normal vs. diseased; retina vs. brain)
uses fluorescent stains/labels to quantify PCR product synthesis
reads fluorescence level at every cycle

37
PCR: Polymerase Chain Reaction
Taq DNA polymeraseIsolated from Thermus aquaticusHeat stableSome have proofreading capability
Recomb DNA 74
PCR: Polymerase Chain Reaction
Key components: template DNA; primers; deoyxribonucleotides; Taq polymerase

38
RT-PCR Analysis of Gene Expression
Agarose gel of RT‐PCR products
cDNA from multiple tissues
RT‐PCR using primers for 6 different genes
Separated on Agarose gel
Stained with ethidium bromide
All are in retina, only F, I, J are retina‐specific (lane #6)
retina retina
Quantitative “real time” PCR
Recomb DNA 76
• PCR with addition of DNA intercalator dye (SYBR green)
• Dye fluoresces only when bound to double stranded DNA
• Detector measures increase in fluorescence after each cycle
• Each cycle doubles the amount of fluorescent PCR product
From: http://eng.bioneer.com/products/GeneExpression/qPCRArrayService-detection.aspxhttp://www.thermoscientificbio.com/qpcr-master-mixes-and-assays/dynamo-hs-sybr-green-qpcr-kit/

39
Recomb DNA 77
Can also start with RNA and generate cDNA for template
Analyze by PCR or amplify targets for cloning
PCR products are often cloned into plasmids
Basic Molecular Methods: Restriction Enzymes
Bacterial enzymes
Normal function : defend against phage infection
Recognizes and cuts (digests) specific DNA sequences
e.g., EcoRI (from E. coli)
5’GAATTC 3’ 5’G AATTC 3’ 3’CTTAAG 5’ 3’CTTAA G 5’
Recognition sequences methylated in the bacterial genome to
prevent DNA digestion by restriction enzymes made by the cell
Wide range of enzymes with different target recognition sites
Commercially available

40
Cloning Using Restriction Enzymes
Cut plasmid and insert DNA with same restriction enzyme
Generate compatible “sticky ends”
Hybridize DNA fragments
Bacterial DNA ligase repairs nicked ends
Grow plasmid in E. coli
Isolation of Plasmid Clones

41
Vector DNA
= Insert (cloned) DNA
Looking for DNA Insert in Plasmid Clones
•Restriction Enzyme digest
•Gel electrophoresis
•Fluorescent stainEthidium bromide = DNA intercalator

42
Recomb DNA 83
DNA SequencingChain Termination Method (Sanger Method)
Components:
• DNA template (genomic DNA, cDNA)
• ONE primer for target
• dNTPs
• ddNTPs (each with radioactive or fluorescent label)
• Taq DNA polymerase
• Amplify by PCR– linear amplification with single primer!
• Each cycle, after a ddNTP is incorporated, DNA synthesis stops
• Generates a collection of all possible DNA fragments with ddNTP at3’ end
• Run gel to size fractionate
• Read sequence by size of each PCR product: speed and distance ofmigration determined by size
Recomb DNA 84
Di-deoxy sequencing method
Di-deoxyNTP (ddATP; ddCTP ddGTP ddTTP)
• Lacks 3’-OH group
• Cannot form a phosphodiester bond with
the next nucleotide in the growing chain
• DNA synthesis is terminated
Video of sequencing:https://www.youtube.com/watch?v=jFCD8Q6qSTM
DNA SequencingChain Termination Method (Sanger Method)

43
• Each ddNTP (ddATP, ddTTP, ddCTP, ddGTP) is labeled with differentfluorescent molecule
• Generates all possible fragments with each labeled at one end• Shown: DNA Fragments generated by di-deoxy sequencing using ddATP
with fluorescent tag ( )
Primer
DNA SequencingChain Termination Method (Sanger Method)
Recomb DNA 86
Automated Chain Termination Sequencing
• Automated sequencer uses electrophoresis that is performed in agel that is inside a thin capillary tube
• Reads fluorescent signal as each product passes detector
• Electropherogram (below) shows fluorescence intensity for eachPCR product
• Each peak shows a fragment with a specific ddNTP at the end
• Read sequence from left to right (small to large fragments)

44
“Next Generation” Sequencing (massively parallel sequencing)for genome and transcriptome
analysis
Recomb DNA 87
Goldman &. Domschke (2014) Making sense of deep sequencingDOI: http://dx.doi.org/10.1017/S1461145714000789 1717-1725
Analyzes all DNA or cDNA in the sample simultaneously
Each fragment 100-500 bp
Total DNA sample is tagged with an adaptor at each end
DNA is attached to “flow cell” or other substrate
The adapter hybridizes to primer for limited PCR to amplify each molecule to generate ‘spot’
Recomb DNA 88
• Uses 4 terminator NTPs that aremodified to stop elongation (likeddNTP)
• Allows addition of one baseadded at a time
• Treat entire flow cell with mix ofall 4 terminators
• Record fluorescence of everyspot on entire flow cell(photograph) after each base isadded
• Chemical treatment removesfluorescent tag and reversesterminator
• Add another batch of the 4terminated
• Repeat
Informative Video:http://www.yourgenome.org/video/sequencing-at-speed

45
Mapping RNA seq data to genome
Recomb DNA 89
• Number of aligned fragments shows relative abundance of differenttranscripts
• Can identify alternative splicing (variable abundance of differentexons)
Recomb DNA 90
Analysis of DNA, mRNA and Protein
Gene (DNA)—Southern blot
Transcription (RNA)-northern blot
Translation (protein)-western blot
General strategy:• Isolate appropriate cellular
component
• Size fractionate by gelelectrophoresis
• (electrical current drivesmolecules through gel)
• Detect using labeled probes(piece of gene of interest;antibody against protein)
• Analyze: compare wild-type vs.experimental, disease vs. healthy,etc

46
Applications of Southern blots
• Identification of a single gene in a pool of DNA fragments
• Detection of specific DNA sequences in a genome
• Studying genomic deletions, duplications, and mutations that cause various diseases
• Genotyping (e.g. transgenic mice)
• Detection of genetic diseases and cancers, such as monoclonal leukemia and sickle cell mutations
• DNA fingerprinting and forensic tests such as paternity testing and sex determination
Recomb DNA 91
Applications of northern blots
• Determine actual size of transcript(s) and reveal presence of alternative splicing in a single gene
• Gene expression studies – e.g. to observe overexpression of cancer-causing genes and gene expression in case of transplant rejection
• In diagnosis of several diseases, e.g. Crohn’s disease
• For detection of viral microRNAs that play key roles in viral infection
• To screen recombinants - by detecting the mRNA formed by the transgene
• Requires that you have RNA from the tissue where it is expressed!!!!
reverse-transcriptase PCR (RT-PCR) can provide a faster and more sensitive alternative (next slide)
Recomb DNA 92

47
Recomb DNA 93
• Start with cDNA from multiple tissues (this was done in mouse!)• RT-PCR using primers for 6 different genes• Separated on agarose gel and stained with ethidium bromide• All are present in retina, but only F, I, J are retina-specific (lane #6)
#6 = retina #6 = retina
Analysis of Gene Expression: RT-PCR of cDNA
Assess differences or changes in RNA expression between:
• Different tissues (e.g.retina vs. brain)
• Normal vs. disease state
• Response totherapy/drug treatments
Microarrays
Recomb DNA 94
Commercially available
• Tens of thousands of spots,each containing many copiesof single stranded DNAprobes
• For gene expression:contains DNA copies (cDNA)for known or predicted genes
• For GWAS/genotyping: allpossible sequence variantsof SNPs
• For mutation analysis: allpossible sequence variantsfor mutations in diseaseassociated genes
http://www.stemcore.ca/services/microarrayshttp://www.ur.umich.edu/0506/Oct17_05/02.shtml
Expected sequence: A T G C A G C C T C T G T C A GVariant (SNP, mutation) A T G C A G C C T T T G T C A G
https://en.wikipedia.org/wiki/DNA_microarray

48
Analysis of Gene Expression: in situ Hybridization
Purpose: Determine the cellular expression of RNA within a tissue
Tissue: frozen or paraffin sections
Probe: anti‐sense RNA (complementary to endogenous RNA)
make by in vitro transcription
labeled with digoxygenin protein‐tagged ribonuclotides
Hybridize probe to RNA in tissue
Antibody detection of protein‐tag; colorimetric detection of antibodies
In situ hybridization to mouse retina
Rhodopsin CRX negative control
Applications of western blots
• Medical diagnosis (some examples listed)
– HIV infection,
– Hepatitis B Virus infections
– Bovine Spongiform Encephalopathy (“mad cow disease”)
– Lyme disease
• Detect presence of protein in a mixture of other proteins
• Determine size and test for post-translational modifications of a protein
• Analysis of protein-protein interactions
• Determine protein stability
• Structure domain analysis
Recomb DNA 96

49
Analysis of Protein Expression: Western Blot
• Total proteins isolated from tissues
• Separated by Poly‐Acrylamide Gel Electrophoresis (PAGE)
• Transferred to nitrocellulose filter
• Incubated with antibody against KLF15 (transcription factor)
• Different isoforms present in different tissues
Recomb DNA 98
Analysis of Protein Expression: Immuno-histochemistry / Immuno-fluorescence
• Purpose: Determine the cellular expression of protein within a tissue
• Tissue: Frozen or paraffin sections
• Probe: Antibodies that recognize specific protein (primary antibody)
• Detection: Tagged antibodies that recognize primary antibody (secondaryantibody)
– Immunohistochemistry: secondary tagged with enzyme; apply substrate for colorimetric detection.
– Immunofluorescence: secondary antibody tagged with fluorescent molecule
Section from fish retina:Stained for 2 different proteins using immuno-fluorescence (red) and immunohistochemistry (brown)

50
Analysis of Gene Function
Expression Vectors (plasmid, virus)Cloned genes are transcribed and translated into protein product
Gene is cloned under control of a strong promoter to drive high levels of expression.• Protein purification/biochemistry• Protein‐Protein interactions• Protein‐DNA interactions• Enzymatic activity
Transgenic animal production (more on this next semester)• Knock‐out mouse: eliminate gene to determine effect of lossof function on normal development/function
• Transgenic Animals: overexpress gene / force expression andanalyze effects
Transgenic Technology Transgenic
general term for genetically modified animal;
typically refers to random gene insertion
Knock-out
transgenic animal with a specific gene deleted/altered in the genome
Knock-in
transgenic animal designed to replace a target gene and with foreign gene
Conditional Knockout
transgenic animal that is engineered to delete/alter a target gene only under certain conditions
John Wilsonhttp://www.bcm.edu/biochem/?PMID=3946
http://www.fao.org/fishery/topic/14796/en
http://www.tetra-fish.com/glofish.aspx

51
Why Create Transgenic Animals?
To produce therapeutic proteins
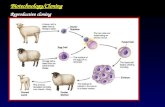
e.g. Transgenic sheep and goats to produce human
proteins in their milk
insulin production
Research tools
Develop models for studying disease
e.g. Lumican knockouts: study corneal structure
MMP‐9 knockouts to study dry eye disease
Rhodopsin knock‐ins to model Retinitis pigmentosa
Replace normal genes with mutated genes to
determine consequences of mutation
Recomb DNA 102
Creating a Transgenic Mouse
Direct injection of DNAcontaining gene of interest into male pro-nucleus of mouse zygote of black mouse
Transplant into surrogate mother with different coat color (albino or brown)
Recover pups with black fur, check for presence of transgene (PCR; Southern)
• Not targeted: Insertion sites are random
• Transgene is typically inserted as multiple end to end copies
• Can induce mutations in unknown genes
• Does not remove existing gene

52
Recomb DNA 103
Generating Targeted Gene Knock-out
Replace original gene with engineered gene • homologous recombination is a rare event• much more difficult, time consuming• can take up to a year or more until you have mice to analyze
Uses cellular DNA repair mechanisms to replace endogenous gene with the engineered targeting construct
Transfect DNA construct into 108 embryonic stem cells
• 5-30 % of cells don’t take up any DNA
• In cells that take up the DNA the gene is not replaced:
• Most frequent: transgene DNA not incorporated into genome
• Others: transgene is randomly insertions
• ~1 cell in 100 million will have homologous recombination event(so, for 1 experiment you may only recover ~10 cells!)
Recomb DNA 104http://sgugenetics.pbworks.com/w/page/24704977/Creation%20of%20a%20%22Knockout%20Mouse%22
1. Embryonic Stem cells (ES cells) fromblasocyst of brown mouse
2. Clone targeting construct• contains mouse DNA from targeted region• engineered DNA containing desired
mutation/insert• genes for drug selection =
HSV-tk (thymidine kinase)neoR (neomycin resistance)

53
Recomb DNA 105
3. Introduce targeting construct into ES cells
4. Possible outcomes:• no DNA in cell frequent not useful• DNA randomly inserted moderate frequency not useful
• homologous recombination: normal gene replaced with targetingconstruct
this is a RARE event, but is the desired result
5. Use drug selection to obtain the rare ES cell with the correctly targeted gene:
• Cells with any of the engineered DNA construct will have neoR and survive
• Only cells with homologous recombination will lack HSV-TK and survive
Recomb DNA 106http://sgugenetics.pbworks.com/w/page/24704977/Creation%20of%20a%20%22Knockout%20Mouse%22
6. Inject ES cells with targeted gene (from brown mouse) intoblastocysts from white mouse
7. Transplant into female host
8. Pups will be chimeras (brown and white)

54
Recomb DNA 107
Chimeras contain mixture of cells with and without transgene
Somatic cells containing transgene: not passed to next generation
Germline cells containing transgene: passed to next generation
Chimeras
Mythical Chimera
Chimeric mouse
chimera.clubhertzog.com/images/chimera.png
http://intramural.nimh.nih.gov/tgc/photogallery.html
Recomb DNA 108http://sgugenetics.pbworks.com/w/page/24704977/Creation%20of%20a%20%22Knockout%20Mouse%22
Breed chimeras to normal mice
Genotype pups to identify mice with targeted gene
Breed to generate homozygous mice
Analyze effects of gene knockout or gene replacement on survival, development, pathology, etc.