04 Med Shunu Biol 2013
Transcript of 04 Med Shunu Biol 2013
-
7/27/2019 04 Med Shunu Biol 2013
1/37
Genu expression
DNS replication
Topic 4
-
7/27/2019 04 Med Shunu Biol 2013
2/37
-
7/27/2019 04 Med Shunu Biol 2013
3/37
Role of RNA and protein synthesis in a
cell
-
7/27/2019 04 Med Shunu Biol 2013
4/37
-
7/27/2019 04 Med Shunu Biol 2013
5/37
-
7/27/2019 04 Med Shunu Biol 2013
6/37
Actin
Nuclear myosin
Transcription
factor
Polymerase I
-
7/27/2019 04 Med Shunu Biol 2013
7/37
-
7/27/2019 04 Med Shunu Biol 2013
8/37
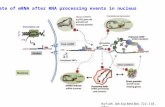
Eukaryotic pre-mRNA processing
5' cap addition
5' capA 5' cap(also termed an RNA cap, an RNA 7-
methylguanosinecap, or an RNA m7G cap) is a modified
guanine nucleotide that has been added to the "front" or 5'
endof a eukaryotic messenger RNA shortly after the start of
transcription. Its presence is critical for recognition by the
ribosomeand protection from RNases.Splicing
Splicing is the process by which pre-mRNA is modified to
remove stretches of non-coding sequences called introns; the
stretches that remain include protein-coding sequences andare called exons. Splicing is usually performed by an RNA-
protein complex called the spliceosome, but some RNA
molecules are also capable of catalyzing their own splicing
(see ribozymes).
http://en.wikipedia.org/
http://en.wikipedia.org/wiki/5'_caphttp://en.wikipedia.org/wiki/7-methylguanosinehttp://en.wikipedia.org/wiki/7-methylguanosinehttp://en.wikipedia.org/wiki/5'_endhttp://en.wikipedia.org/wiki/5'_endhttp://en.wikipedia.org/wiki/Ribosomehttp://en.wikipedia.org/wiki/RNasehttp://en.wikipedia.org/wiki/Intronhttp://en.wikipedia.org/wiki/Exonhttp://en.wikipedia.org/wiki/Spliceosomehttp://en.wikipedia.org/wiki/Ribozymehttp://en.wikipedia.org/wiki/Ribozymehttp://en.wikipedia.org/wiki/Spliceosomehttp://en.wikipedia.org/wiki/Exonhttp://en.wikipedia.org/wiki/Intronhttp://en.wikipedia.org/wiki/RNasehttp://en.wikipedia.org/wiki/Ribosomehttp://en.wikipedia.org/wiki/5'_endhttp://en.wikipedia.org/wiki/5'_endhttp://en.wikipedia.org/wiki/7-methylguanosinehttp://en.wikipedia.org/wiki/7-methylguanosinehttp://en.wikipedia.org/wiki/7-methylguanosinehttp://en.wikipedia.org/wiki/5'_cap -
7/27/2019 04 Med Shunu Biol 2013
9/37
Editing
mRNA can be edited, changing the nucleotide composition of thatmRNA. An example in humans is the apolipoprotein BmRNA,
which is edited in some tissues, but not others. The editing createsan early stop codon, which, upon translation, produces a shorter
protein.
Polyadenylation
Polyadenylation is the covalent linkage of a polyadenylyl moiety to a
messenger RNA molecule. In eukaryotic organisms all messengerRNA (mRNA) molecules are polyadenylated at the 3' end. The
poly(A) tailand the protein bound to it aid in protecting mRNAfrom degradation by exonucleases.
Polyadenylation occurs during and immediately after transcription of
DNA into RNA. After transcription has been terminated, themRNA chain is cleaved through the action of an endonucleasecomplex associated with RNA polymerase. After the mRNA has
been cleaved, around 250 adenosine residues are added to the free3' end at the cleavage site. This reaction is catalyzed by
polyadenylate polymerase.
http://en.wikipedia.org/
http://en.wikipedia.org/wiki/RNA_editinghttp://en.wikipedia.org/wiki/Apolipoprotein_Bhttp://en.wikipedia.org/wiki/Messenger_RNAhttp://en.wikipedia.org/wiki/Messenger_RNAhttp://en.wikipedia.org/wiki/Apolipoprotein_Bhttp://en.wikipedia.org/wiki/RNA_editing -
7/27/2019 04 Med Shunu Biol 2013
10/37http://en.wikipedia.org/
-
7/27/2019 04 Med Shunu Biol 2013
11/37
-
7/27/2019 04 Med Shunu Biol 2013
12/37
RNA processing and splicing
http://www.youtube.com/watch?v=YjWuVrzvZYA
http://www.youtube.com/watch?v=FVuAwBGw_pQ
http://www.youtube.com/watch?v=YjWuVrzvZYAhttp://www.youtube.com/watch?v=FVuAwBGw_pQhttp://www.youtube.com/watch?v=FVuAwBGw_pQhttp://www.youtube.com/watch?v=YjWuVrzvZYA -
7/27/2019 04 Med Shunu Biol 2013
13/37
-
7/27/2019 04 Med Shunu Biol 2013
14/37
-
7/27/2019 04 Med Shunu Biol 2013
15/37
Ribosomes in the cytoplasm and polysomes
attached to the endoplasmic reticulul (ER).
-
7/27/2019 04 Med Shunu Biol 2013
16/37
http://www.youtube.com/watch?v=NJxobgkPEAo
Ribosomes
urnls "Nature " 9/27/2001
Ribosomas prvietoans virziens
http://www.youtube.com/watch?v=NJxobgkPEAohttp://www.youtube.com/watch?v=NJxobgkPEAohttp://www.youtube.com/watch?v=NJxobgkPEAo -
7/27/2019 04 Med Shunu Biol 2013
17/37
-
7/27/2019 04 Med Shunu Biol 2013
18/37
-
7/27/2019 04 Med Shunu Biol 2013
19/37
4. stage:
5. stage:
6. stage:
7. stage:
How many phosphates arenecessary to produce asigle polypeptide (100amino acids)
-
7/27/2019 04 Med Shunu Biol 2013
20/37
Proteins after translation
http://www.sumanasinc.com/webcontent/animations/content/lifecycleprotein.html
http://www.sumanasinc.com/webcontent/animations/content/lifecycleprotein.htmlhttp://www.sumanasinc.com/webcontent/animations/content/lifecycleprotein.html -
7/27/2019 04 Med Shunu Biol 2013
21/37
-
7/27/2019 04 Med Shunu Biol 2013
22/37
-
7/27/2019 04 Med Shunu Biol 2013
23/37
Proteins of the secretory pathway are translocated into the endoplasmic reticulum(ER) lumen co-translationally through proteinaceous channels in the ERmembrane called translocons. b | In the extremely crowded, calcium-rich,oxidizing environment of the ER lumen, resident chaperones like BiP, calnexinand protein disulphide isomerase (PDI) serve to facilitate the proper folding of the
nascent protein by preventing its aggregation, monitoring the processing of thehighly branched glycans, and forming disulphide bonds to stabilize the folded
protein. c | Once correctly folded and modified, the protein will exit the ERthrough the formation of transport vesicles and move on through the secretory
pathway. d | If the ER quality-control system deems that the protein is malfoldedor unable to fold, it will be targeted for retrotranslocation to the cytosol and
degraded by the 26S proteasome. e | Changes in the ER environment shift thebalance from normal folding to improper folding (thicker arrow), leading to theaccumulation of unfolded proteins in the ER. This activates three ER-stresssensors IRE1, PKR-like ER kinase (PERK) and ATF6 which initiate theunfolded protein response. SRP, signal-recognition particle.
S
-
7/27/2019 04 Med Shunu Biol 2013
24/37
Secretory proteins
signalpeptide
Ribosome
Ribosome
mRNA
mRNA
receptorpeptidase
translocator
-
7/27/2019 04 Med Shunu Biol 2013
25/37
-
7/27/2019 04 Med Shunu Biol 2013
26/37
ER LUMENCYTOSOL
glucose
mannose
N-acetylglucosamine
-
7/27/2019 04 Med Shunu Biol 2013
27/37
ER and turnover of ER resident proteins
-
7/27/2019 04 Med Shunu Biol 2013
28/37
Golgi complex and N-linked glycosylation
-
7/27/2019 04 Med Shunu Biol 2013
29/37
-
7/27/2019 04 Med Shunu Biol 2013
30/37
-
7/27/2019 04 Med Shunu Biol 2013
31/37
-
7/27/2019 04 Med Shunu Biol 2013
32/37
-
7/27/2019 04 Med Shunu Biol 2013
33/37
-
7/27/2019 04 Med Shunu Biol 2013
34/37
-
7/27/2019 04 Med Shunu Biol 2013
35/37
-
7/27/2019 04 Med Shunu Biol 2013
36/37
-
7/27/2019 04 Med Shunu Biol 2013
37/37